
MiR-195-5p suppresses the proliferation, migration, and invasion of gallbladder cancer cells by targeting FOSL1 and regulating the Wnt/β-catenin pathway
Introduction
Gallbladder carcinoma (GBC) is a malignant tumor originating from the gallbladder epithelium and is one of the most common and aggressive tumors (1). The clinical symptoms of patients with GBC are not obvious and the onset is relatively insidious, enabling cancer cells to spread widely in the early stage through direct infiltration, lymphatic metastasis, and blood circulation (2,3). Radical surgery is the most effective treatment for GBC, but most patients have already entered the middle and late stages at the time of diagnosis and have missed the optimal period for surgery (4). Moreover, the efficacy of adjuvant therapy, such as chemotherapy and radiotherapy, in patients with advanced GBC is short-lived, and chemotherapy and radiotherapy resistance is common (5). Therefore, the overall prognosis of GBC is very poor, with a 5-year survival rate of less than 5% (6). Despite major efforts to identify tumor-promoting and tumor suppressor genes in GBC over the past few decades, few independent biomarkers have been identified that can be routinely used in clinical (7,8). Therefore, a new potential GBC marker to address this issue is eagerly sought.
MicroRNAs (miRNAs) are endogenous, single-stranded small RNAs of 20–24 nt in length that are highly conserved and regulate gene expression by degrading target messenger RNAs (mRNAs) or inhibiting their translation (9,10). Accumulating evidence suggests that mature miRNAs play important roles in various physiological and pathological processes, such as cell proliferation, migration, invasion, and signal transduction (11). Previous studies have found that multiple miRNAs are abnormally expressed in GBC tissues and are associated with factors such as GBC metastasis, tumor size, and lymph node metastasis, including the upregulation of miR-181b-5p expression or the downregulation of miR-204, miR-551b-3p, and miR-30a-5p expression (12-15). It has been reported that miR-195-5p can act as a tumor suppressor in various cancers (16-18); however, whether miR-195-5p can affect the migration and invasion of GBC and thereby inhibit its progression is still unclear.
Fos-like antigen-1 (FOSL1), a member of the AP1 complex, heterodimerizes with members of the JUN family for efficient transcriptional activity (19). The mRNA and protein expression of FOSL1 is upregulated in multiple tumor types, and this molecule exerts oncogenic effects by regulating various cellular processes, such as proliferation, differentiation, invasion, epithelial-mesenchymal transition (EMT), and drug resistance (20). The Wnt/β-catenin signaling pathway is one of the key signaling pathways in cancer, and studies have shown that FOSL1 can participate in the regulation of the Wnt/β-catenin signaling pathway and promote the progression of colorectal cancer (21,22). However, the roles of FOSL1 and the Wnt/β-catenin signaling pathway in GBC are still unclear. The biological roles of miR-195-5p/FOSL1 regulatory axis and Wnt/β-catenin signaling pathway in gallbladder cancer have not been reported in other literatures.
In this study, the importance of the role of miR-195-5p, FOSL1, and the Wnt/β-catenin signaling pathway in the development of GBC was explored through bioinformatics methods combined with in vitro and in vivo experiments. We aimed to yield new insights for molecular targeted therapy. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3685/rc).
Methods
Bioinformatics analysis
We obtained the miRNA dataset GSE104165 and the mRNA dataset GSE74048 by searching the Gene Expression Omnibus database (GEO; https://www.ncbi.nlm.nih.gov/geo/) and used the Limma package of R language software to perform differential gene analysis (23). The screening criteria for differentially expressed miRNAs were |log fold change (FC)|>2 and adjusted P-value <0.05, and the screening criteria for differentially expressed mRNAs were |log FC|>2 and P-value <0.05. FunRich software (http://www.funrich.org/) was used to predict the target genes of differentially expressed miRNAs, and Perl (http://www.perl.org/) software was used to intersect the predicted target genes of differentially expressed miRNAs with the differentially expressed mRNAs in the dataset to construct a miRNA-mRNA regulatory network. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Human intrahepatic biliary epithelial cells (HIBEpiCs) and the GBC cell lines GBC-SD and NOZ were provided by Jenniobio Biotechnology Co. (Guangzhou, China). Cells were cultured in Dulbecco’s modified Eagle medium [DMEM; fetal bovine serum (FBS) 10% + penicillin (100 U/mL) + streptomycin (100 mg/mL), Gibco, Waltham, MA, USA] at 37 ℃ and 5% CO2. Fresh culture medium was regularly replaced every 2–3 days, and cells were routinely digested and passaged when confluence reached 80%.
Cell treatment and transfection
The miR-195-5p mimic, negative control (NC) mimic, miR-195-5p inhibitor, and NC inhibitor were purchased from GenePharma (Shanghai, China). The miR-195-5p mimic (40 nM), NC mimic (40 nM), miR-195-5p inhibitor (100 nM), or NC inhibitor (100 nM) was transfected into GBC-SD or NOZ cells with Lipofectamine® 3000 (Thermo Fisher Scientific, Waltham, MA, USA).
Lentiviral human FOSL1-targeting short hairpin RNA (shRNA) was designed via the Thermo Fisher Scientific website (https://rnaidesigner.thermofisher.com/rnaiexpress/sort.do). The target shRNAs against the human FOSL1 gene (GenBank accession NM_001300844.2) for RNAi were sh-FOSL1 #1: 5'-GCTGTGGTCAAGGAGGTAAGT-3' and sh-FOSL1 #2: 5'- GCACGAAGACGTTGATATAGC -3'; PLKO.1 blank lentivector was used as a NC. For overexpression of FOSL1, the complementary DNA (cDNA) of a fragment encoding the full-length human FOSL1 open reading frame (ORF) sequence was amplified by polymerase chain reaction (PCR) and then cloned into a pSin lentiviral expression vector (oe-FOSL1) to obtain a recombinant lentivirus. An overexpression miR195-5p lentiviral plasmid was constructed using pCDH vectors (Lv-miR195-5p), and empty pCDH vectors acted as NCs (Lv-NC) (GuangZhou Yingxin Biological Technology, Guangdong, China).
The cells were treated with 2 µg/mL puromycin for 7 days to obtain stable cell lines after 72 hours of infection. Real-time quantitative PCR (qRT-PCR) was used to verify the expression levels of FOSL1 and miR195-5p, and western blotting was used to verify the expression levels of FOSL1.
In the groups treated with dimethyl sulfoxide (DMSO) or PNU74654 [MedChemExpress (MCE), Monmouth Junction, NJ, USA], 15 µL DMSO or 129.8 µM PNU74654 were added to the culture medium. After 48 hours of treatment, the cells were harvested and used for subsequent assays.
RNA isolation and quantitation
The miRNA was extracted from cultured cells using a miRNA Purification Kit, and total RNA was extracted from cultured cells using an Ultrapure RNA Kit (both from CWBIO, Beijing, China). The RNA samples were then reverse transcribed into cDNA with a miRNA cDNA synthesis kit and a HiFiScript cDNA synthesis kit (CWBIO, China). We performed qRT-PCR with the miRNA qPCR Assay Kit and UltraSYBR Mixture (CWBIO, China) using a LightCycler 480 System (Roche, Indianapolis, IN, USA). The expression of miRNA was quantified with the U6 control, and the mRNA levels were quantified with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA expression as an endogenous control. The relative levels of gene expression were calculated based on the 2-△△Ct method. The primers used were designed and synthesized by Sangon Biotech (Shanghai, China) (Table 1).
Table 1
| Target gene | Primer (5'-3') |
|---|---|
| miR-29c-3p | F: CGCGCTAGCACCATTTGAAATCGGTT |
| miR-125b-5p | F: CGCTCCCTGAGACCCTAACTTGTGA |
| miR-144-3p | F: CCGCGCGCGTACAGTATAGATGATGT |
| miR-145-5p | F: CCGTCCAGTTTTCCCAGGAATCCCT |
| miR-195-5p | F: CGCGCTAGCAGCACAGAAATATTGGC |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| FOSL1 | F: ACTGGAAGATGAGAAATCTGGG |
| R: GGGAAAGGGAGATACAAGGTAC | |
| ESRRG | F: CACATAGAAGATGTTGAAGCCG |
| R: TGATGTTGTAGAAATGCTGCAC | |
| CEP55 | F: ATGCAGGCATGTACTTTAGACT |
| R: AAGCTGTTTCAAGGATTCCAAC | |
| CBLC | F: GTGAGTATCTACCAGTTCCACG |
| R: CCCCTTTAGGAGTCTCACTTTC | |
| EFNA1 | F: TGAGGACTACACCATACATGTG |
| R: CTGCCACAGAGTGATCTTCATA | |
| GAPDH | F: GCACCGTCAAGGCTGAGAAC |
| R: TGGTGAAGACGCCAGTGGA |
Western blot analysis
Proteins were extracted from cells using precooled radioimmunoprecipitation assay (RIPA) buffer (CWBIO, China) with protease and phosphatase inhibitor cocktails (CWBIO, China). The protein concentration was determined using a bicinchoninic acid (BCA) protein quantification kit (Beyotime Biotechnology, Shanghai, China). Protein samples (20 µg) were separated through polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were blocked in 5% nonfat dry milk for 1 hour and then incubated with primary antibodies to β-tubulin (#66240‐1‐Ig; 1:20000), FOSL1 (ab252421; 1:1000), β-catenin (ab32572; 1:1000), CCND1 (ab134175; 1:10000), or c-Myc (ab32072; 1:1000) overnight at 4 ℃. The membranes were then incubated with the corresponding secondary rabbit or mouse antibody tagged by horseradish peroxidase (CWBIO, China). Protein bands were visualized using enhanced chemiluminescence reagents (ECL; Biosharp Life Sciences, Anhui, China) and photographed using a ChemiDoc Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Densitometric analysis was performed using ImageJ 1.53c software (National Institutes of Health, Bethesda, MD, USA) with normalization to the expression of β-tubulin.
Dual-luciferase reporter assay
The free online tool TargetScan (http://www.targetscan.org/vert_72/) was used to predict the binding sites of miR-195-5p to FOSL1. The FOSL1 3'-untranslated region (3'UTR) containing the wild-type binding site predicted by miR-195-5p was amplified and cloned into the pmirGLO vector (FOSL1-WT) (Promega Corp., Madison, WI, USA). The FOSL1 3'UTR containing the binding site of the miR-195-5p mutation was obtained by site-directed mutagenesis (the possible binding sequence of miR-195-5p and FOSL1 3'UTR was deleted). The mutant sequence was amplified and cloned into pmirGLO vector to construct the FOSL1-MUT plasmid. At 48 hours after transfection with NC mimic or miR-195-5p mimic, luciferase activity was detected using a dual-luciferase reporter assay system (Solarbio Corp., Beijing, China) and normalized to Renilla activity.
Cell counting kit-8 assay
The GBC-SD and NOZ cells were cultured in a 96-well plate at a density of 5,000 cells/well for 0, 24, 48, and 72 hours. The cultured medium in each well was exchanged with fresh medium containing 10% Cell Counting Kit-8 (CCK-8; Absin Bioscience, Inc., Shanghai, China) and incubated at 37 ℃ for 2 hours. The optical density (OD) values of the medium were measured at 450 nm using a MultiskanTM FC Microplate Photometer (Thermo Fisher Scientific, USA).
Cell wound healing assays and transwell assays
For the cell wound healing assay, GBC cells were cultured in 24-well plates to generate a confluent monolayer (2×105 cells per well) and then scratched with a 200 µL sterile pipette tip. After being gently washed twice with phosphate-buffered saline (PBS), the wounded cell monolayer was incubated with FBS-free medium at 37 ℃. The scratch wounds were photographed with an inverted phase-contrast microscope (Olympus, Hamburg, Germany) at 0, 24, and 48 hours after scratching and then assessed by ImageJ software. We set the initial scratch width to 100%, and the results were expressed as the percentage of wound closure.
Cell invasion assays were performed using transwell chambers (8.0 µm pore size; Corning, Corning, NY, USA). Matrigel [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA] was thawed at 4 ℃, and 50 µL of thawed Matrigel was added to the chambers. After incubation at room temperature for 5 hours, a total of 100 µL of serum-free medium containing 5×104 cells was added to the upper chamber, and 600 µL of 10% FBS culture medium was added to the lower chamber. After incubation at 37 ℃ and 5% CO2 for 24 hours, noninvaded cells on the top of the transwell were removed, and the invading cells were fixed with 4% paraformaldehyde and stained with crystal violet staining solution (0.1% w/v, Beyotime Biotechnology, China). Photographs were taken randomly from 5 regions of each membrane, and the number of invading cells was expressed as the average number of cells in each region. All assays were independently repeated 3 times.
Cell cycle analysis
Cell cycle distribution was measured by flow cytometry. After the appropriate treatments, cell cycle distribution was determined using a cell cycle staining kit (MultiSciences, Hangzhou, China) according to the manufacturer’s protocol. The GBC cells were collected and fixed with 70% cold ethanol. Next, propidium iodide (PI) staining solution with RNase A was added and incubated in the dark at 37 ℃ for 30 minutes. The stained samples were tested by a CytoFLEX flow cytometer (Beckman Coulter, Carlsbad, CA, USA), and the results were analyzed by ModFit (Verity Software House, Topsham, ME, USA).
Immunofluorescent staining
The GBC cells were seeded in 15 mm confocal dishes (NEST Biotech., Jiangsu, China) and incubated for 24 hours before staining. The cells were fixed in 4% paraformaldehyde and then permeabilized in 0.1% Triton X-100 at room temperature. After being blocked for 1 h with phosphate-buffered saline/Tween (PBST) containing 10% goat serum, the cells were incubated with β-catenin primary antibodies (ab32572; 1:200) overnight at 4 ℃. After 3 washes with PBST, a cocktail of fluorescence-conjugated secondary antibody (ab150078, 1:100, Abcam, Cambridge, MA, USA) was added to the cells. Then, the cell nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI; C0065, Solarbio Corp., China), and the cells were photographed under a confocal microscope (LSM880 with Fast Airyscan; ZEISS, Oberkochen, Germany).
Immunohistochemistry
Immunohistochemistry was performed to observe the expression of FOSL1 (ab252421; 1:500), Ki67 (A2094; 1:2,000), β-catenin (ab32572; 1:500), CCND1 (ab134175; 1:500), and c-Myc (ab32072; 1:500). Sections were incubated with a primary antibody at 4 ℃ overnight, washed with PBS 3 times, and incubated with secondary antibody at room temperature for 1 hour. Thereafter, the sections were incubated with DAB (AR1022, Boster Company, China) for immunohistochemistry staining. After counterstaining with hematoxylin, the slides were prepared for microscopic (Olympus, Germany) evaluation.
Tumor xenograft model
In this study, 10 Male BALB/c nude mice aged 4–6 weeks (16–18 g) were purchased from Beijing Vital River Laboratory Animal Technology Co. (Beijing, China) and randomly divided into 2 groups (Lv-miR-195-5p and Lv-NC) with 5 mice in each group. The animals were kept in the same environment [specific-pathogen-free (SPF) animal laboratory] to eliminate interference.
Equal numbers (3×106 cells) of Lv-miR-195-5p and Lv-NC GBC-SD cells were subcutaneously injected into BALB/c nude mice. Xenograft mouse models were kept for 36 days and tumor volumes were recorded every 4 days after injection. Tumor volumes were measured using a standard caliper and were calculated with the following formula: V=(L×W2)/2 (L, the longest tumor axis; W, the shortest tumor axis). At the end of the experiments, all nude mice were sacrificed, and the xenograft tumors were weighted and collected for immunohistochemistry. A protocol was prepared before the study without registration. Experiments were performed under a project license (No. IACUC-2021031202) granted by the Animal Ethical and Welfare Committee at the Sixth Affiliated Hospital of Sun Yat-sen University, in compliance with institutional guidelines for the care and use of laboratory animals.
Statistical analysis
All statistical analyses were performed using SPSS 26.0 statistical software (IBM Corp., Armonk, NY, USA). All data are presented as the mean ± standard deviation (SD) of at least 3 independent experiments. An unpaired 2-tailed Student’s t-test was used for comparative analysis between the 2 groups. One-way analysis of variance (ANOVA) was used to compare multiple groups, followed by Tukey’s post-hoc test. A P-value less than 0.05 was considered significant.
Results
MiRNA-mRNA coregulatory network in GBC
We retrieved and obtained relevant datasets for GBC samples from the GEO database and then performed bioinformatics analysis on these datasets (Figure 1A). First, we used R language to analyze the GBC miRNA dataset GSE104165 to determine the differentially expressed miRNAs (|log FC|>2, adjusted P<0.05) between GBC and normal gallbladder tissues and generated a heatmap (Figure 1B) and a volcano map (Figure 1C). Moreover, we used FunRich software to predict the target genes of all differentially expressed miRNAs. Next, we analyzed the GBC mRNA dataset GSE74048, screened out the differentially expressed mRNAs between GBC and normal gallbladder tissues (|log FC|>2, P<0.05), and generated a heatmap (Figure 1D) with a volcano plot (Figure 1E). Then, the target genes predicted for the miRNAs were further intersected with the mRNAs screened from the GSE74048 dataset to obtain the possible miRNA-mRNA regulatory network of GBC (Figure 1F). Finally, the top 5 miRNA-mRNA regulatory axes ranked by |log FC| were selected for experimental verification, and their roles and mechanisms in the progression of GBC were further studied (Table 2). We used qRT-PCR to confirm that both miR-145-5p and miR-195-5p levels were significantly downregulated in 2 GBC cell lines, GBC-SD and NOZ, compared with those in HIBEpiCs (Figure 1G). Therefore, we further identified the expression of potential target genes of miR-145-5p and miR-195-5p and confirmed that FOSL1 was elevated in GBC cells compared with HIBEpiCs (Figure 1H).
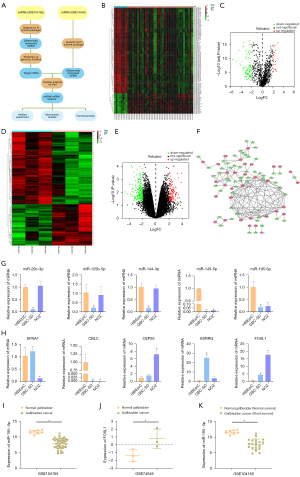
Table 2
| miRNA | Target | mRNA | miRNA LogFC | mRNA LogFC |
|---|---|---|---|---|
| hsa-miR-145-5p | Target | EFNA1 | −4.852705757 | 2.113080402 |
| hsa-miR-125b-5p | Target | ACHE | −4.057789761 | 2.117387521 |
| ESRRG | −4.057789761 | 2.733009085 | ||
| SGOL1 | −4.057789761 | 2.245136804 | ||
| VAV3 | −4.057789761 | 2.504915294 | ||
| hsa-miR-29c-3p | Target | MYBL2 | −3.696319794 | 2.020220853 |
| hsa-miR-144-3p | Target | CEP55 | −3.675952033 | 2.771505528 |
| PMAIP1 | −3.675952033 | 2.208247395 | ||
| STIL | −3.675952033 | 2.388986806 | ||
| hsa-miR-195-5p | Target | CBLC | −3.672802065 | 2.16986385 |
| CEP55 | −3.672802065 | 2.771505528 | ||
| ESRRG | −3.672802065 | 2.733009085 | ||
| FOSL1 | −3.672802065 | 2.155042439 |
miRNA, microRNA; mRNA, messenger RNA; GBC, gallbladder cancer; FC, fold change.
Moreover, we analyzed the expression of miR-195-5p and FOSL1 in their respective GEO datasets. The expression of miR-195-5p in GBC was significantly lower than that in normal gallbladder tissue (Figure 1I) and the expression of FOSL1 in GBC was higher than that in normal gallbladder tissue (Figure 1J). Further analysis of the relationship between miR-195-5p and patient survival prognosis in the data set showed that the decreased expression of miR-195-5p was associated with poor patient prognosis (Figure 1K).
The relationship between the miR-195-5p/FOSL1 regulatory axis and GBC was preliminarily determined.
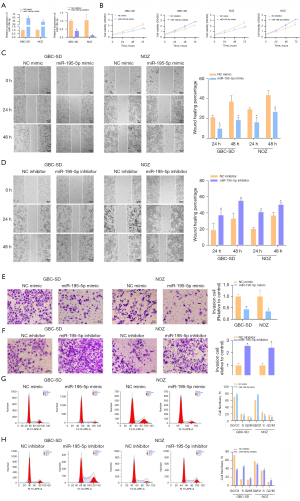
Overexpression of miR-195-5p inhibits the proliferation, migration, and invasion of GBC cells
To study the biological function of miR-195-5p in GBC cells, we first used qRT-PCR to detect the transfection efficiency of miR-195-5p. As shown, the expression of miR-195-5p was significantly upregulated in both cell lines after transfection of the miR-195-5p mimic, whereas the miR-195-5p inhibitor had the opposite effect (Figure 2A). The results of CCK-8 assays and statistical analysis showed that the proliferation of the GBC-SD and NOZ cells overexpressing miR-195-5p was significantly lower than that of the control group, and the proliferation of the miR inhibitor-transfected cells was significantly enhanced (Figure 2B). The effect of miR-195-5p on GBC-SD and NOZ cell migration was examined using wound healing assays. After statistical analysis, overexpression of miR-195-5p was shown to significantly inhibit the movement of GBC-SD and NOZ cells, and the migration of the miR inhibitor-transfected group was significantly higher than that of the control group (Figure 2C,2D). Transwell assays were used to detect the effect of miR-195-5p on the invasion of GBC-SD and NOZ cells. The statistical results showed that the number of GBC-SD and NOZ cells with high expression of miR-195-5p in the transwells was significantly lower than that of the control cells. The number of cells transfected with the miR inhibitor was significantly higher than that of the control cells, and the difference was significant compared with that of the control group (Figure 2E,2F). Flow cytometry and statistical analysis showed that miR-195-5p overexpression resulted in GBC-SD and NOZ cell cycle arrest in the G0/G1 phase and a significant reduction in cells in the S phase. Transfection of the miR inhibitor reduced the cells in the G0/G1 phase and increased those in the S phase, and the difference was significant (Figure 2G,2H). In conclusion, miR-195-5p has a tumor suppressor effect in GBC.
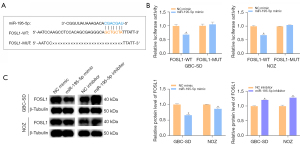
FOSL1 is the direct target of miR-195-5p in GBC
We used the online bioinformatics tool TargetScan to find the possible binding sequence of miR-195-5p and FOSL1 and constructed the pmirGLO dual luciferase FOSL1-WT and FOSL1-MUT reporter plasmids (Figure 3A). The results showed that the miR-195-5p mimic decreased the luciferase activity of the FOSL1-WT-transfected GBC cells but did not affect the luciferase activity of the FOSL1-MUT-transfected GBC cells (Figure 3B). In addition, our western blot results showed that the transfected miR-195-5p mimic significantly downregulated the protein level of FOSL1 in GBC cells, and correspondingly, the changes after transfection with the miR-195-5p inhibitor were reversed (Figure 3C). Thus, FOSL1 is a direct target of miR-195-5p in GBC cells and negatively correlated with miR-195-5p.
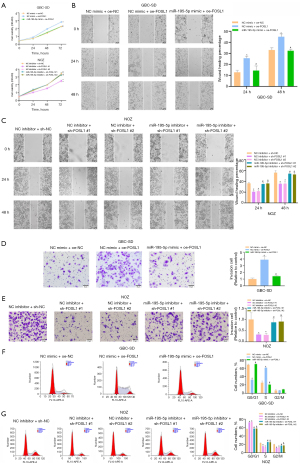
MiR-195-5p targets FOSL1 to inhibit the proliferation, migration, and invasion of GBC cells
To further explore the biological role of the miR-195-5p/FOSL1 axis in GBC, we established the GBC cell line GBC-SD with stable overexpression of FOSL1 (oe-FOSL1) by lentivirus and constructed a NOZ GBC cell line with stable knockdown of FOSL1 (sh-FOSL1 #1, sh-FOSL1 #2) by lentivirus. The transfection efficiency was verified by qRT-PCR and western blots (Figure S1A,S1B). We separately treated the FOSL1-overexpressing GBC-SD and FOSL1 knockdown NOZ cells and examined their proliferation (Figure 4A), migration (Figure 4B,4C), invasion (Figure 4D,4E), and cell cycle (Figure 4F,4G). The results and statistical analysis showed that the proliferation, migration, and invasion of GBC cells were enhanced after overexpression of FOSL1 (P<0.05). Compared with those of the GBC cells in the NC-mimic+oe-FOSL1 group, the proliferation, migration, and invasion of the GBC cells in the miR-195-5p mimic + oe-FOSL1 group were decreased (P<0.05). After overexpression of FOSL1, the proportion of cells in the G0/G1 phase was significantly decreased, and the proportion of cells in the S phase was significantly increased (P<0.05). Compared with that of the GBC cells in the NC-mimic + oe-FOSL1 group, the proportion of cells in the G0/G1 phase in the miR-195-5p mimic + oe-FOSL1 group was significantly increased, and the proportion of cells in the S phase was significantly decreased (P<0.05). Correspondingly, after knockdown of FOSL1, the proliferation, migration, and invasion of GBC cells were weakened, the proportion of cells in the G0/G1 phase was significantly increased, and the proportion of cells in the S phase was significantly decreased (P<0.05), but the miR-195-5p inhibitor could reverse these changes (P<0.05). Therefore, miR-195-5p targeted the regulation of FOSL1 to inhibit the proliferation, migration, and invasion of GBC cells.
MiR-195-5p regulates the Wnt/β-catenin signaling pathway by targeting FOSL1
In this study, after transfection of the miR-195-5p mimic and miR-195-5p inhibitor, western blotting was used to detect changes in Wnt/β-catenin signaling pathway proteins in GBC cells. The results showed that the miR-195-5p mimic could reduce the protein expression levels of β-catenin, c-Myc, and CCND1 (Figure 5), and the miR-195-5p inhibitor could increase their expression levels (Figure 5B). We also studied the mechanism by which the miR-195-5p/FOSL1 axis regulates the Wnt/β-catenin signaling pathway by rescue experiments. Western blot and immunofluorescence results indicated that overexpression of FOSL1 could increase the nuclear expression of β-catenin and activate the Wnt/β-catenin signaling pathway, and this effect could be reversed by the miR-195-5p mimic (Figure 5C,5E). Correspondingly, knockdown of FOSL1 reduced the nuclear expression of β-catenin and inhibited the activation of the Wnt/β-catenin signaling pathway, and the miR-195-5p inhibitor reversed this effect (Figure 5D,5F). These results suggest that miR-195-5p regulates the Wnt/β-catenin signaling pathway by targeting FOSL1.
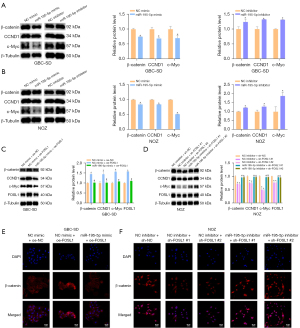
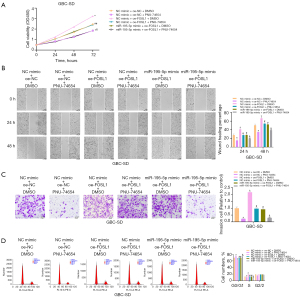
We treated the FOSL1-overexpressing GBC-SD cells with miR-195-5p or PNU74654 and examined their proliferation (Figure 6A), migration (Figure 6B), invasion (Figure 6C), and cell cycle (Figure 6D). The results and statistical analysis showed that the Wnt/β-catenin signaling pathway involved in miR-195-5p/FOSL1 axis mediated the proliferation, migration, and invasion of GBC cells.
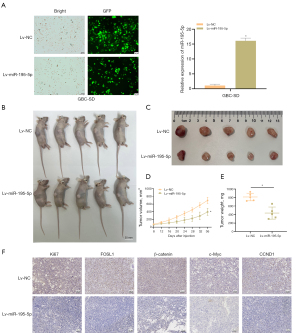
Upregulation of miR-195-5p expression suppresses xenograft tumor growth in BALB/c-nu mice
To confirm whether miR-195-5p affects the tumorigenesis of GBC in vivo, we established the GBC cell line GBC-SD with stable overexpression of miR-195-5p (Lv-miR-195-5p) mediated by lentivirus and the blank control (Lv-NC), and the transfection efficiency was detected by fluorescence microscopy and qRT-PCR (Figure 7A). We subcutaneously injected Lv-miR-195-5p and Lv-NC GBC cells to establish a xenograft model, and after the 8th day, the subcutaneous tumor volume was measured every 4 days. On day 36, the mice were sacrificed, and the tumors were photographed and weighed (Figure 7B-7E). Compared with those of the control group, the tumor volume and weight of the mice in the Lv-miR-195-5p group decreased significantly (P<0.05). Immunohistochemistry was used to verify the effect of miR-195-5p overexpression on tumor proliferation and the expression of Ki67, FOSL1, β-catenin, c-Myc, and CCND1. The expression of Ki67, FOSL1, β-catenin, c-Myc, and CCND1 was lower in the tumors of the Lv-miR-195-5p group mice (Figure 7F). These results indicate that miR-195-5p inhibits the growth of xenograft tumors in BALB/c-nu mice.
Discussion
Characteristically, GBC has an insidious onset and a high degree of malignancy, is prone to metastasis, has limited advanced treatment options, and has a 5-year survival rate of less than 5% (24). Therefore, identification of highly specific markers for GBC is urgently needed.
In recent years, miRNAs have been confirmed to play an important role in the progression of a variety of solid tumors, are closely related to survival and prognosis, and are considered one of the most valuable tumor markers (25). Recent studies have shown a strong relationship between miRNAs and gallbladder cancer. MiR-365 can inhibit the progression of gallbladder cancer and predict the prognosis of gallbladder cancer patients (26); circulating miR-141 is a potential biomarker for the diagnosis, prognosis and therapeutic target of gallbladder cancer (27); miR-4733-5p promotes gallbladder cancer progression via directly targeting kruppel like factor 7 (28). An important mechanism of miRNAs in tumors is to bind to the complementary sequence of the 3'UTR of specific target mRNAs to degrade or inhibit these molecules, thereby regulating biological behaviors such as tumor proliferation, migration, and invasion. The miRNA-mRNA coregulatory networks are ubiquitous in organisms and are closely related to cancer. For example, in glioma, miR-29a-5p alters cell proliferation, migration, and invasion by targeting DHRS4 (29); miR-29c-3p targeting CCNA2 regulates migration, invasion, and the cell cycle in esophageal cancer (30). Therefore, determining the important miRNA-mRNA coregulatory network in cancer and further elucidating its mechanism are key to cancer research. In this study, the abnormally expressed miRNAs and mRNAs in GBC patients were screened out by bioinformatics analysis, and the miRNA-mRNA coregulatory network was constructed according to their interactions. We confirmed that the expression of miR-195-5p was decreased and the expression of FOSL1 was increased in 2 GBC cell lines, preliminarily indicating that this miRNA-mRNA coregulatory network may play an important role in the progression of GBC.
MiR-195-5p is a multitarget regulator involved in various aspects of cancer cell behavior (31). It can regulate the cell cycle and inhibit the growth of leukemia cells; its expression is increased and is an important prognostic factor (32). Furthermore, stable expression of miR-195-5p in a mouse xenograft model of ovarian cancer significantly reduced tumor growth, increased tumor doubling time, and improved overall survival (33). In the present study, after we treated GBC cells with the miR-195-5p mimic, the proliferation, migration, and invasion were significantly reduced, and cell cycle arrest in the G0/G1 phase was confirmed by flow cytometry. The opposite results were obtained after treating GBC cells with a miR-195-5p inhibitor. These results confirmed the important role of miR-195-5p in the progression of GBC.
As a member of the AP1 complex, FOSL1 acts as an oncogenic factor in tumor development. FOSL1 is involved in a complex network of interactions with miRNAs, EMT-inducing transcription factors (EMT-TFs), and cytokines (34). In this study, the overexpression of miR-195-5p resulted in inhibition of the target gene FOSL1, as shown by luciferase reporter and western blot analyses. Recent studies have shown that FOSL1 is a major regulator in head and neck squamous cell carcinoma (HNSCC), suggesting that FOSL1 is a potential prognostic biomarker in HNSCC (35). In our study, overexpression of FOSL1 in GBC-SD enhanced its proliferation, migration, and invasion and promoted the transition of the cell cycle of GBC cells from the G0/G1 phase to the S phase. The opposite biological changes were observed in FOSL1 knockdown NOZ cells, which confirmed the cancer-promoting effect of FOSL1 in GBC. Moreover, we demonstrated in further rescue experiments that the oncogenic effect of FOSL1 in GBC is directly mediated by miR-195-5p.
Abnormalities in the Wnt/β-catenin signaling pathway can promote cancer stem cell renewal, cell proliferation, and differentiation, thus playing a key role in tumorigenesis and treatment response (36). Activation of the Wnt/β-catenin signaling pathway can affect changes in the downstream molecules CCND1 and c-Myc (37). Our experiments showed that miR-195-5p targets FOSL1 to regulate the Wnt/β-catenin signaling pathway in GBC, thereby promoting changes in GBC cell proliferation, migration, and invasion. The downstream molecule CCND1 of this signaling pathway is a marker protein of the cell cycle transition from the G0/G1 phase to the S phase (38). When the cell cycle is blocked in G0/G1 phase, the expression of CCND1 decreases, which is consistent with our experimental results. In nude mouse tumorigenesis experiments, we also showed that miR-195-5p regulates FOSL1 and the Wnt/β-catenin signaling pathway in GBC, thereby affecting tumor growth.
This study still had certain limitations. The interaction mechanism between FOSL1 and the Wnt/β-catenin signaling pathway is not fully understood; moreover, the diagnostic value of miR-195-5p in GBC still needs to be verified with a large number of clinical specimens. Therefore, we hope to conduct more basic experiments in the future and collect more specimens of GBC cases and follow-up data to address these issues.
Conclusions
The overexpression of miR-195-5p inhibits the proliferation, invasion, and migration of GBC cells through direct targeting of FOSL1 and regulation of the Wnt/β-catenin signaling pathway, which provides new ideas for the diagnosis and treatment of GBC.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (Nos. 30671987 and 8180102403), PhD Start-up Fund of Natural Science Foundation of Guangdong Province (No. 2015A030310377), and National Key Clinical Discipline.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3685/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3685/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3685/coif). All authors report that this work was supported by grants from the National Natural Science Foundation of China (Nos. 30671987 and 8180102403), PhD Start-up Fund of Natural Science Foundation of Guangdong Province (No. 2015A030310377) and National Key Clinical Discipline. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license (No. IACUC-2021031202) granted by the Animal Ethical and Welfare Committee at the Sixth Affiliated Hospital of Sun Yat-sen University, in compliance with institutional guidelines for the care and use of laboratory animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Apodaca-Rueda M, Cazzo E, De-Carvalho RB, et al. Prevalence of gallbladder cancer In patients submitted to cholecystectomy: experience of the University Hospital, Faculty of Medical Sciences, State University of Campinas - UNICAMP. Rev Col Bras Cir 2017;44:252-6. [Crossref] [PubMed]
- García P, Lamarca A, Díaz J, et al. Current and New Biomarkers for Early Detection, Prognostic Stratification, and Management of Gallbladder Cancer Patients. Cancers (Basel) 2020;12:3670. [Crossref] [PubMed]
- Kim BH, Kwon J, Chie EK, et al. Adjuvant Chemoradiotherapy is Associated with Improved Survival for Patients with Resected Gallbladder Carcinoma: A Systematic Review and Meta-analysis. Ann Surg Oncol 2018;25:255-64. [Crossref] [PubMed]
- Valle JW, Kelley RK, Nervi B, et al. Biliary tract cancer. Lancet 2021;397:428-44. [Crossref] [PubMed]
- Lazcano-Ponce EC, Miquel JF, Muñoz N, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin 2001;51:349-64. [Crossref] [PubMed]
- Sharma A, Sharma KL, Gupta A, et al. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol 2017;23:3978-98. [Crossref] [PubMed]
- Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872-6. [Crossref] [PubMed]
- Mishra SK, Kumari N, Krishnani N. Molecular pathogenesis of gallbladder cancer: An update. Mutat Res 2019;816-818:111674. [Crossref] [PubMed]
- Chen L, Heikkinen L, Wang C, et al. Trends in the development of miRNA bioinformatics tools. Brief Bioinform 2019;20:1836-52. [Crossref] [PubMed]
- Catalanotto C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int J Mol Sci 2016;17:1712. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Qin Y, Zheng Y, Huang C, et al. Downregulation of miR-181b-5p Inhibits the Viability, Migration, and Glycolysis of Gallbladder Cancer by Upregulating PDHX Under Hypoxia. Front Oncol 2021;11:683725. [Crossref] [PubMed]
- Zhang B, Cui H, Sun Y, et al. Up-regulation of miR-204 inhibits proliferation, invasion and apoptosis of gallbladder cancer cells by targeting Notch2. Aging (Albany NY) 2021;13:2941-58. [Crossref] [PubMed]
- Ji T, Gao L, Yu Z. Tumor-suppressive microRNA-551b-3p targets H6PD to inhibit gallbladder cancer progression. Cancer Gene Ther 2021;28:693-705. [Crossref] [PubMed]
- Ye YY, Mei JW, Xiang SS, et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis 2018;9:410. [Crossref] [PubMed]
- Chae DK, Park J, Cho M, et al. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-β receptor I ubiquitination. Mol Oncol 2019;13:2663-78. [Crossref] [PubMed]
- Sun M, Song H, Wang S, et al. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J Hematol Oncol 2017;10:79. [Crossref] [PubMed]
- Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer 2019;18:4. [Crossref] [PubMed]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 2003;3:859-68. [Crossref] [PubMed]
- Talotta F, Casalino L, Verde P. The nuclear oncoprotein Fra-1: a transcription factor knocking on therapeutic applications' door. Oncogene 2020;39:4491-506. [Crossref] [PubMed]
- Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol 2020;13:165. [Crossref] [PubMed]
- Liu Y, Yue M, Li Z. FOSL1 promotes tumorigenesis in colorectal carcinoma by mediating the FBXL2/Wnt/β-catenin axis via Smurf1. Pharmacol Res 2021;165:105405. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Rakić M, Patrlj L, Kopljar M, et al. Gallbladder cancer. Hepatobiliary Surg Nutr 2014;3:221-6. [PubMed]
- Orso F, Quirico L, Dettori D, et al. Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin Cancer Biol 2020;60:214-24. [Crossref] [PubMed]
- Jiang ZB, Ma BQ, Feng Z, et al. miR-365 inhibits the progression of gallbladder carcinoma and predicts the prognosis of Gallbladder carcinoma patients. Cell Cycle 2021;20:308-19. [Crossref] [PubMed]
- Yang G, Lu Z, Meng F, et al. Circulating miR-141 as a potential biomarker for diagnosis, prognosis and therapeutic targets in gallbladder cancer. Sci Rep 2022;12:10072. [Crossref] [PubMed]
- Hu X, Zhang J, Bu J, et al. MiR-4733-5p promotes gallbladder carcinoma progression via directly targeting kruppel like factor 7. Bioengineered 2022;13:10691-706. [Crossref] [PubMed]
- Dai Y, Chen Z, Zhao W, et al. miR-29a-5p Regulates the Proliferation, Invasion, and Migration of Gliomas by Targeting DHRS4. Front Oncol 2020;10:1772. [Crossref] [PubMed]
- Wang H, Fu L, Wei D, et al. MiR-29c-3p Suppresses the Migration, Invasion and Cell Cycle in Esophageal Carcinoma via CCNA2/p53 Axis. Front Bioeng Biotechnol 2020;8:75. [Crossref] [PubMed]
- Yu W, Liang X, Li X, et al. MicroRNA-195: a review of its role in cancers. Onco Targets Ther 2018;11:7109-23. [Crossref] [PubMed]
- Boldrin E, Gaffo E, Niedermayer A, et al. MicroRNA-497/195 is tumor suppressive and cooperates with CDKN2A/B in pediatric acute lymphoblastic leukemia. Blood 2021;138:1953-65. [Crossref] [PubMed]
- Rao G, Dwivedi SKD, Zhang Y, et al. MicroRNA-195 controls MICU1 expression and tumor growth in ovarian cancer. EMBO Rep 2020;21:e48483. [Crossref] [PubMed]
- Dhillon AS, Tulchinsky E. FRA-1 as a driver of tumour heterogeneity: a nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene 2015;34:4421-8. [Crossref] [PubMed]
- Zhang M, Hoyle RG, Ma Z, et al. FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol Ther 2021;29:2583-600. [Crossref] [PubMed]
- Yu F, Yu C, Li F, et al. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther 2021;6:307. [Crossref] [PubMed]
- Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther 2022;7:3. [Crossref] [PubMed]
- Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol 2013;14:518-28. [Crossref] [PubMed]
(English Language Editor: J. Jones)


