
MicroRNA-668-3p regulates oxidative stress and cell damage induced by Aβ1-42 by targeting the OXR1/p53-p21 axis
Introduction
Alzheimer’s disease (AD) is a degenerative disease that is common in the elderly and is characterized by progressive memory loss and dementia (1,2). Due to increased longevity globally, the treatment of AD has become paramount for public healthcare systems (3,4). Although some drugs for AD have been approved for marketing (5,6), AD still has an unclear pathogenesis and no effective cure.
Increasing evidence shows that oxidative stress may be a key factor in the pathogenesis of AD (7,8). Excessive production of oxidative stress and free radicals may lead to neuronal death, thereby promoting the loss of cognitive ability (9,10). Additionally, amyloid β (Aβ) is closely related to the severity of cognitive decline, and Aβ1-42 is the main factor driving AD development (11,12). Studies have shown that oxidative stress is the earliest change in AD development, and it also participates in the subsequent stages of AD and is therefore the most important mechanism in AD pathogenesis (13-15). Chen et al. (16) identified that oxidative stress participates in AD progression by promoting Aβ deposition.
MicroRNAs (miRNAs) are important factors that affect the occurrence and development of various diseases such as breast cancer (17) and Parkinson’s disease (18). Studies have suggested that various physiological and pathological processes of the nervous system are also affected by miRNAs, which have been identified as potential biomarkers of AD (19-21). For example, miR-668-3p has an important regulatory effect on a variety of cells, such as liver cancer cells and cardiomyocytes, in terms of biological function (22,23). However, to the best of our knowledge, there has been no relevant research on the role of miR-668-3p in AD.
MiRNAs can affect the cell cycle, proliferation, differentiation, migration, invasion, and apoptosis by regulating downstream target genes and signaling pathways (24-28). Oxidation resistance 1 (OXR1) plays a role in antioxidative stress and has an effect on neurodegenerative diseases (29,30). Increased OXR1 expression can effectively protect against oxidative stress-induced cell damage (31,32).
Various factors that are upregulated in cancer cells to maintain growth and survival are downregulated in AD, leading to neurodegeneration. AD develops when the growth and antistress response of aging neurons are weakened, and the regulation of cell death and maintenance mechanisms are modified (33). Proteins p21 and p53 are important tumor-suppressor factors and have been the focus of various studies related to AD. The occurrence and development of AD can be promoted by p21 and p53 through excessive enhancement of their levels and continued maintenance of tau phosphorylation. Therefore, p21 and p53 have been identified as potential biomarkers for AD (34-36).
This study aimed to explore whether miR-668-3p acts downstream of OXR1 to affect p53-p21 signaling and mediate the oxidative stress induced by Aβ1-42, thereby alleviating AD progression. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3598/rc).
Methods
Animals
Six 120-days-old female C57BL/6 and C57BL/6 (AD model) specific pathogen-free mice were obtained from the Guangdong Medical Experimental Animal Center (License No. SCXK 2018-0002; Guangzhou, China) and placed in a polystyrene cage in a room with constant temperature (23±2 ℃) and humidity (45%±15%) conditions, a 12-hour light/dark cycle, and ad libitum access to standard food and water. After a week of adaptive feeding, the mice were euthanized by intraperitoneal injection of sodium pentobarbital (150 mg/kg body weight). Thereafter, the brain tissue was aseptically removed, the cerebral cortex was carefully opened, the hippocampus was exposed, and the surrounding tissues of the hippocampus were separated using ophthalmic scissors and placed into Hank’s balanced salt solution (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). A protocol was prepared before the study without registration. Animal experiments were approved by the Animal Care and Use Committee of Guangxi Medical University (approval No. 2017JJB10090) and were conducted in accordance with the National Institutes of Health guidelines for the care and use of animals.
Hematoxylin-eosin (HE) staining
The brain tissue was placed in 10% formalin for 48 hours, then rinsed with water for 24 hours to remove formalin from the tissue. After dehydration with a gradient concentration of ethanol, the brain tissue was soaked in xylene I for 40 minutes to render it transparent and then embedded in paraffin wax. The tissue was cut into 4-µm-thick slices, fixed on slides, dried, and then stained with HE staining solution (Solarbio) according to the manufacturer’s instructions. The slices were soaked in xylene, then in a gradient concentration of ethanol, and subsequently in hematoxylin, and then sealed with resin. After drying, the structure of the hippocampus was observed under a light microscope.
Determination of reactive oxygen species (ROS)
The brain tissue was cut to a size of 1 mm3 and washed twice with phosphate-buffered saline (PBS) to remove tissue debris and blood. Thereafter, the tissue was rapidly frozen with liquid nitrogen, homogenized using a homogenizer, centrifuged (1,500 ×g, 4 ℃, 20 minutes), and the supernatant was collected and resuspended in prechilled PBS. The obtained single-cell suspension was incubated at 37 ℃ for 1 hour with 10 µM 2',7'-dichlorodihydrofluorescein diacetate, centrifuged (1,000 ×g, 10 minutes), and washed twice with precooled PBS. The fluorescence intensity of dichlorofluorescein was measured at 488 nm using a multifunctional microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Enzyme linked immunosorbent assay (ELISA)
The cell supernatant or serum of each subgroup was collected after centrifugation (1,000 ×g, 10 minutes) to analyze the levels of nitric oxide (NO), malondialdehyde (MDA), superoxide dismutase (SOD), and glutathione peroxidase (GSH). The following kits were used according to the manufacturer’s instructions: NO content assay kit (BC1475, Solarbio), MDA assay kit (BC0025, Solarbio), GSH assay kit (BC1195, Solarbio), and SOD activity detection kit (BC5160, Solarbio).
Microarray raw dataset analysis using the Gene Expression Omnibus (GEO)
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Raw gene expression data (GSE157239_RAW.tar and GSE150696_RAW.tar) were downloaded from the GEO website (http://www.ncbi.nlm.nih.gov/geo) and divided into 4 groups: control case miRNAs (GSM4759791, GSM4759794, GSM4759796), AD case miRNAs (GSM4759793, GSM4759797, GSM4759804), control case mRNAs (GSM4556860, GSM4556861, GSM4556862), and AD case mRNAs (GSM4556851, GSM4556852, GSM4556853). The Affymetrix Transcriptome Analysis Console (version 4.0) was used to analyze the Affymetrix Human Genome U133 Plus 2.0 Array to generate a heatmap of differentially expressed miRNAs and mRNAs. Differentially expressed RNAs were considered significant at a P value of ≥0.01 and a log2 (fold change, FC) >1.5. Sequences of the 3 small interfering RNAs (siRNAs) targeting OXR1, the miR-668-3p mimic or inhibitor, and the negative control (NC) are shown in Table S1.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay
Total RNA from neuronal stem cells (NSCs) was extracted using TRIzolTM reagent (Thermo Fisher Scientific). After addition of 200 µL of chloroform and vigorous shaking and mixing for 30 seconds, the sample was left to stand for 10 minutes. Next, the upper aqueous phase was collected after centrifugation (14,000 ×g, 15 minutes) to obtain the RNA pellets. The cDNA was reverse transcribed using the PrimeScript RT reagent kit (Takara, Dalian, China) according to the manufacturer’s recommendations. RT-qPCR analysis was performed using the SYBR® Premix Ex Taq™ II kit (Takara Bio, Shiga, Japan) and Applied Biosystems® 7500 Real-Time PCR System (CXF96; Bio-Rad, Hercules, CA, USA). The PCR conditions were as follows: 50 ℃ for 2 minutes, 95 ℃ for 2 minutes, followed by 40 cycles of 95 ℃ for 15 seconds and 60 ℃ for 32 seconds. The primers used for RT-qPCR analysis are listed in Table S2. MiR-668-3p and OXR1 RNA levels were normalized to those of U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated using the 2−ΔΔCq method (37).
Immunohistochemistry (IHC) and immunofluorescence (IF) assay
Brain tissue sections were incubated in 3% H2O2 to quench the endogenous peroxidase activity. The sections were then incubated overnight at 4 ℃ with a rabbit polyclonal anti-OXR1 antibody (1:50 for IHC, 2 µg/mL for IF; ab251774; Abcam, Cambridge, UK), followed by incubation for 30 minutes at 37 ℃ with an appropriate horseradish peroxidase (HRP)-conjugated secondary goat anti-rabbit antibody (for IHC) or goat anti-rabbit Alexa Fluor® 647 (1:50 for IF, ab190565, Abcam), followed by staining with 3,3'-diaminobenzidine or 4',6-diamidino-2-phenylindole (for IF).
Western blot assay
After the NSCs were lysed with lysis buffer (Solarbio), protein concentrations in the extracts were determined using a bicinchoninic acid (BCA) protein assay kit (Solarbio) according to the manufacturer’s instructions. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (10%) was used to separate denatured proteins (20 µg), which were then transferred to polyvinylidene fluoride membranes (MilliporeSigma, Burlington, MA, USA). The membranes were rinsed with 10% tris-buffered saline (TBS)-Tween-20 (Solarbio), blocked with 5% bovine serum albumin (Solarbio), and incubated overnight at 4 ℃ with antibodies against OXR1 (0.4 µg/mL, ab251774, Abcam), p53 (1:10,000, ab32389, Abcam), and p21 (1:1,000, ab109520, Abcam). Membranes were then incubated at 25 ℃ for 2 hours with a secondary antibody (goat anti-rabbit, 1:10,000, ab205718, Abcam). GAPDH (1:10,000, ab181602, Abcam) was used as the loading control.
Isolation and culture of NSCs
The brain tissue was cut into pieces of approximately 1 mm3, digested with 0.125% trypsin, and incubated at 37 ℃ for 20 minutes, with gentle shaking 2–3 times during the incubation period. The stop solution and filter were added to prepare the cell suspension, which was then centrifuged (1,000 ×g, 10 minutes, 4 ℃). The pellet was resuspended in complete medium and inoculated into a culture flask (37 ℃, 5% CO2). The NSCs were cultured for 6 hours, and the medium was then replaced with serum-free medium. After 3 days, Ara-c (Sigma-Aldrich, St. Louis, MO, USA) was added for 24 hours to inhibit the proliferation of nonneuronal cells. The medium was changed every 3 days for 21 days during the cell experiments.
Identification of NSCs
NSCs (5×104) were inoculated into a 6-well plate (Corning, Corning, NY, USA) for 24 hours on round coverslips. After 3 rinses with PBS, Triton X-100 was added for 2 hours. NSCs were incubated with bovine serum albumin for 30 minutes, followed by overnight incubation with anti-MAP2 (1:500, ab254264, Abcam) and anti-β III tubulin (3 µg/mL, ab18207, Abcam) antibodies at 4 ℃ in the dark. After rinsing with PBS 3 times, the cells were incubated at 37 ℃ for 1 hour with goat anti-rabbit Alexa Fluor® 488 (1:200, ab150077, Abcam) or 647 labeled secondary antibodies. Finally, the cells were mounted using a gold antifade mounting medium containing 4',6-diamidino-2-phenylindole (ProLong™, Thermo Fisher Scientific).
Dual luciferase assays
NSCs were transfected with 0.5 µg each of a miR-668-3p mimic/inhibitor, 1 µg each of the plasmids expressing wild-type (WT) or mutant (mut) OXR1, and 0.05 µg of pRL-SV40 reporter vector plasmid. After NSCs were incubated for 48 h, luciferase activity was measured at 490 nm (dual luciferase reporter assay system; Promega Corp., Fitchburg, WI, USA). The ratio of firefly to Renilla is normalized with firefly values.
Cell proliferation assay
NSCs were seeded in 96-well plates at 2.5×103 cells/well and cultured. According to the manufacturer’s instructions, optical density values were measured at time nodes of 0, 24, 48, and 72 h with an enzyme-labeled instrument (Thermo Fisher Scientific) using the Cell Counting Kit-8 (Solarbio) assay.
Flow cytometry (FCM) analysis of apoptosis, cell cycle, and ROS of NSCs
For analysis of apoptosis, A dose of 5 µL of Annexin V-Fluorescein Isothiocyanate (FITC; BD Biosciences, Franklin Lakes, NJ, USA) and 10 µL of propidium iodide (PI; BD Biosciences) were used in combination with the following conditions: dark, 15 min, 25 ℃. Then rinsed twice with PBS (Gibco, Thermo Fisher Scientific).
For cell cycle analysis, NSCs were washed twice with cold PBS and incubated at 37 ℃ for 30 minutes in the dark with PI (400 µL) and RNase (100 µL). The PI signal was detected using an FCM analyzer (BD Biosciences) and the ratio of G1, S and G2 phases in each group of cells was calculated.
For ROS analysis, NSCs were incubated with 2',7'-dichlorodihydrofluorescein diacetate was added to each group of cells at a dose of 1.0 µM and incubated under following conditions: 15 min and 37 ℃. Subsequently, Cells were then gently washed twice with PBS and via FCM to detect ROS level.
Statistical analysis
All experiments were performed in triplicate. Data are expressed as the mean ± standard deviation. All statistical analyses were performed using SPSS version 19.0 statistical analysis package (SPSS Corp., Armonk, NY, USA). Statistical analysis was performed using one-way analysis of variance with a Bonferroni post-hoc test. The means of the 2 groups were analyzed using the Student’s t-test. Differences were considered significant at P<0.05.
Results
Oxidative stress is elevated in AD
Analysis of HE staining showed the nerve cells in the brain tissue of the healthy control group to be neatly arranged and round, with a complete cell structure, clear cell membrane and cell nucleus, and no visible swelling or necrosis. Multiple nerve cells in the brain tissue of AD mice were extremely disordered and irregular in size and shape. The number of nerve cells decreased sharply, and the cell structure was blurry (Figure 1A). Additionally, the results suggested that the levels of ROS, MDA, and NO were increased, whereas the levels of SOD and GSH were decreased in the brain tissue of AD mice compared to those of the healthy control group (Figure 1B-1F). These results confirmed that oxidative stress levels were elevated in the AD mice.
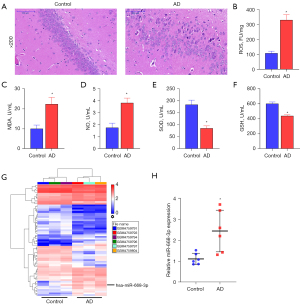
MiRNA screening
To explore the molecular mechanisms that affect AD progression, we analyzed the GSE157239 dataset using GEO. Compared to healthy individuals, in patients with AD, there were 67 dysregulated miRNAs (Figure 1G), of which 31 were upregulated and 36 were downregulated (Table S3). Subsequently, we selected miR-668-3p with the highest FC value as a potential therapeutic target for AD and explored its role in AD pathogenesis. In our subsequent RT-qPCR analysis, we found that miR-668-3p expression in the brain tissue of AD mice was significantly increased (Figure 1H). Therefore, miR-668-3p may be the key to AD development, which was consistent with the results of the GEO data analysis.
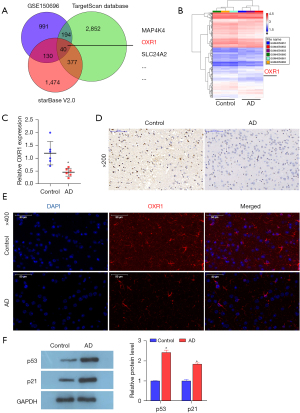
MiR-668-3p binds to OXR1
As miRNAs can regulate downstream signaling pathways by binding to mRNA, we conducted a joint analysis using the TargetScan and starBase databases and the mRNA mined from GEO (dataset GSE150696) (Figure 2A). We speculated that 40 mRNAs had the potential to bind to miR-668-3p. Genes with potential binding sites for miR-668-3p are listed in Table S4. Among these mRNAs, the role of the conserved gene family antioxidant gene 1 (OXR1) in protecting species from oxidative stress has been widely confirmed (29,32). Accordingly, OXR1 was downregulated in AD patients in GEO dataset GSE150696 (Figure 2B). However, there have been no studies discussing the role of OXR1 in AD. Subsequent analysis of RT-qPCR, IHC, and IF results revealed that the RNA and protein levels of OXR1 decreased in the brain tissue of AD mice compared to those in the healthy control group (Figure 2C-2E), indicating that reduced OXR1 levels correlated with AD progression. Additionally, multiple studies have shown that the p53 signaling pathway may be one of the main signaling pathways that affect AD development (38-40). We analyzed p53 signaling pathway-related proteins p53 and p21 by western blotting, and the results revealed that the protein levels of p53 and p21 increased significantly in AD mice (Figure 2F). Therefore, we believe that miR-668-3p may regulate p53 signal transduction by targeting the 3'UTR of OXR1 and promoting the oxidative stress response in brain tissue.
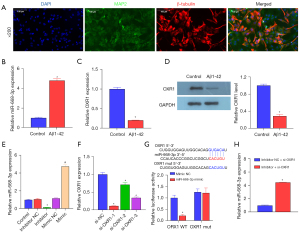
MiR-668-3p inhibitor relieves the Aβ1-42-induced effects by regulating OXR1 expression
To explore the mechanism of the miR-668-3p/OXR1/p53-p21 axis, we extracted and isolated murine hippocampal NSCs. The IF results showed that the expression of MAP2 and β-tubulin in the isolated and cultured hippocampal neuronal cells were both positive (Figure 3A), indicating that the neuronal cells were successfully isolated. Subsequently, Aβ1-42 was used to induce AD in mouse neuronal cells. RT-qPCR and western blotting results suggested that compared to the healthy control group, after Aβ1-42 induction, the expression of miR-668-3p increased (Figure 3B), and the mRNA and OXR1 protein levels decreased (Figure 3C,3D). These results were consistent with those of previous in vivo experiments. We transfected the synthetic miR-668-3p mimic or inhibitor into mouse neuron cells. The results suggested that miR-668-3p RNA levels increased in the mimic group and decreased in the inhibitor group, confirming the effectiveness of the synthetic miR-668-3p mimic or inhibitor (Figure 3E). RT-qPCR results showed that after transfection of the 3 synthesized siRNAs targeting OXR1, the expression of OXR1 decreased to varying degrees. Among them, si-OXR1-1 showed the highest inhibitory efficiency (Figure 3F) and was therefore used to act as an OXR1 antagonist (si-OXR1) for further molecular mechanism research. The results of the dual luciferase assay performed to confirm the direct binding between miR-668-3p and OXR1 showed that in normal hippocampal neuronal cells, compared with the WT OXR1 + miR-668-3p mimic NC group, the fluorescence activity of the miR-668-3p mimic group was significantly decreased. Compared to the mut OXR1 + miR-668-3p inhibitor NC group, the mut OXR1 + miR-668-3p inhibitor group showed no significant difference in fluorescence activity (Figure 3G). RT-qPCR results suggested that suppression of miR-668-3p expression significantly increased the mRNA level of OXR1 (Figure 3H). These results demonstrated that miR-668-3p directly targeted OXR1.
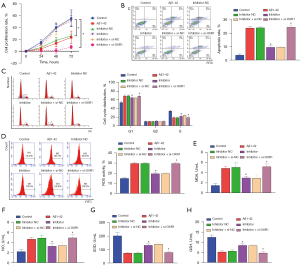
MiR-668-3p negatively regulates OXR1 expression
The results of the rescue experiments showed that Aβ1-42 induced a significant reduction in hippocampal NSC proliferation (Figure 4A), increased apoptosis (Figure 4B) and cell cycle arrest (Figure 4C), and promoted oxidative stress (ROS, MDA, and NO levels increased, and SOD and GSH levels decreased; Figure 4D-4H). The addition of the miR-668-3p inhibitor partially reversed the effect induced by Aβ1-42, but suppression of OXR1 expression prevented the miR-668-3p inhibitor from exerting its effect. These results confirmed the negative regulatory relationship between miR-668-3p and OXR1.
MiR-668-3p regulates the p53 signaling pathway via OXR1
The above in vivo experiments confirmed that the protein levels of p53 and p21 increased significantly in AD, which was verified in vitro using mouse hippocampal NSCs. Tenovin-1 (TEN; activator of the p53 signaling pathway) was cotransfected with the miR-668-3p inhibitor. Western blotting results showed that Aβ1-42 induced a decrease in OXR1 protein levels and an increase in p53/p21 protein levels, and TEN alone enhanced the effect of Aβ1-42 on p53/p21 but had no effect on OXR1 protein levels. The miR-668-3p inhibitor alone reversed the effect of Aβ1-42 on p53 and p21 protein levels by promoting the translation of OXR1, but the effect of the miR-668-3p inhibitor was partially weakened by the combined action of TEN (Figure 5A). Furthermore, the results showed that compared to the healthy control group, Aβ1-42 decreased cell proliferation (Figure 5B) and increased apoptosis (Figure 5C), cell cycle arrest (Figure 5D), and oxidative stress levels (Figure 5E-5I). TEN alone enhanced the effect induced by Aβ1-42, whereas the miR-668-3p inhibitor alone partially reversed the effect of Aβ1-42. Furthermore, the effect of the miR-668-3p inhibitor under the combined action of TEN and Aβ1-42 was significantly inhibited.
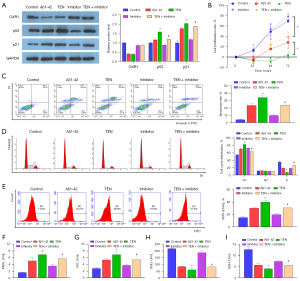
In summary, miR-668-3p suppression was mediated by the p53-p21 pathway and negatively regulated OXR1 expression by targeting the 3'UTR of OXR1. Therefore, miR-668-3p may be a potential therapeutic target for AD.
Discussion
Dysregulated miRNAs, including miR-106b (36), miR-196a (37), and miR-124 (15), are important factors in the development of many degenerative diseases (41,42). For example, recent studies have shown that low levels of miRNAs miR-27a-3p (43) and high levels of miR-483-3p (44) can promote the development of AD. Moreover, miR-342-3p (45) and miR-103a-3p (46) affect the occurrence and subsequent stages of AD. The purpose of our study was to clarify the mechanisms underlying AD development by screening for miRNAs that affect AD progression. Among the murine homologous miRNAs, we found that the FC value of miR-668-3p was the highest. Currently, no follow-up study has investigated miR-668-3p as a therapeutic target for AD. Therefore, we screened miR-668-3p as a potential therapeutic target for AD and explored its role in AD pathogenesis. Subsequently we found that the suppression of miR-668-3p expression had a neuroprotective effect on cell damage induced by AD or Aβ1-42. Furthermore, this neuroprotective effect was related to the inhibition of oxidative stress and neuronal apoptosis as well as the regulation of the miR-668-3p/OXR1/p53-p21 axis.
Studies have shown that AD is characterized by the production of excessive amounts of ROS, MDA, and NO, and inhibition of the secretion of SOD, GSH, and other antioxidant enzymes (47,48), resulting in damage to the hippocampal neurons in the brain (49), which is consistent with our results. These large amounts of indelible ROS can induce damage to cell membranes and mitochondria, leading to NSC apoptosis (50). Additionally, we found that miR-668-3p was upregulated in the GEO data analysis, AD mouse models, and in vitro Aβ1-42-induced NSCs. These results were consistent with those of previous studies (51-53), and we found that in vitro Aβ1-42 induction decreased NSC proliferation and promoted apoptosis, cell cycle arrest, and oxidative stress. By suppressing miR-668-3p expression, the adverse effects of Aβ1-42 induction on cells could be reversed. This finding was similar to that of previous reports (54,55). Therefore, miR-668-3p expression may be the key to AD progression; however, the mechanism of miR-668-3p remains unclear.
It is well known that miRNAs can regulate the transcription and translation of mRNA by binding to the 3'UTR region of mRNA (28). OXR1 is an important protein known to protect cells from oxidative stress (56,57), and it plays a key role in preventing and alleviating neurodegeneration (29). Subsequent RT-qPCR, IHC, and IF experiments confirmed that OXR1 RNA and protein levels were downregulated in vivo and in vitro in the Aβ1-42 cell model of AD mice, a finding similar to that of Jiang et al. in degenerative diseases (Parkinson’s disease) (32). Subsequent luciferase experiments confirmed that miR-668-3p directly targeted the 3'UTR end of OXR1. Thus far, we can speculate that the positive effect of suppressing miR-668-3p expression in cells may have been achieved through OXR1.
A study has shown that various pathways, such as p53, TNF-α, and PI3K/AKT/MTOR, are negatively correlated with cancer in AD (33). We confirmed through rescue experiments that the suppression of OXR1 expression could reverse the effect of the miR-668-3p inhibitor on Aβ1-42 induction. As an activator of p53, the addition of TEN partially reversed the effect of the miR-668-3p inhibitor on Aβ1-42 induction. These results also confirmed the negative correlation between miR-668-3p and OXR1 or p53-p21, in which miR-668-3p regulates p53-p21 signaling by directly targeting OXR1, affecting the proliferation, apoptosis, cell cycle arrest, and oxidative stress levels of NSCs induced by Aβ1-42 in vitro. This study demonstrates that dysregulation of the miR-668-3p/OXR1/p53-p21 pathway is a key mediator of AD pathogenesis, highlights the importance of epigenetics and identifies novel therapeutic targets for AD.
There were some limitations to this study. First, there were no clinical data to support the role of miR-668-3p in patients with AD. Moreover, there are still many mRNAs derived from GEO data mining that have not been analyzed and verified. These are the directions and goals of our future research.
Conclusions
The suppression of miR-668-3p expression had a neuroprotective effect on cell damage induced by AD in vivo and Aβ1-42 in vitro through inhibition of cell cycle arrest, apoptosis, and oxidative stress. The neuroprotective mechanism involved the regulation of the miR-668-3p/OXR1/p53-p21 axis, and miR-668-3p may be a potential AD biomarker.
Acknowledgments
Funding: This work was supported by grants received from the Guangxi Natural Science Foundation for Young Scientists (No. 2017JJB140090z), Guangxi Zhuang Autonomous Region Health Care Commission Self-financing Research Projects (Nos. Z20200096, Z20201417), Guangxi Medicine and Healthy Self-Financing Research Projects (No. Z20180956), and Nanning Qingxiu District Key Research and Development Program (No. 2019041).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3598/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3598/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3598/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. 2017JJB10090) granted by the Animal Care and Use Committee of Guangxi Medical University, in compliance with the National Institutes of Health guidelines for the care and use of animals. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Høgh P. Alzheimer's disease. Ugeskr Laeger 2017; [PubMed]
- Alzheimer's Association. 2016 Alzheimer's disease facts and figures. Alzheimers Dement 2016;12:459-509. [Crossref] [PubMed]
- Weller J, Budson A. Current understanding of Alzheimer's disease diagnosis and treatment. F1000Res 2018;7:eF1000 Faculty Rev-1161.
- Lane CA, Hardy J, Schott JM. Alzheimer's disease. Eur J Neurol 2018;25:59-70. [Crossref] [PubMed]
- Briggs R, Kennelly SP, O'Neill D. Drug treatments in Alzheimer's disease. Clin Med (Lond) 2016;16:247-53. [Crossref] [PubMed]
- Mangialasche F, Solomon A, Winblad B, et al. Alzheimer's disease: clinical trials and drug development. Lancet Neurol 2010;9:702-16. [Crossref] [PubMed]
- Nunomura A, Perry G. RNA and Oxidative Stress in Alzheimer's Disease: Focus on microRNAs. Oxid Med Cell Longev 2020;2020:2638130. [Crossref] [PubMed]
- Tönnies E, Trushina E. Oxidative Stress, Synaptic Dysfunction, and Alzheimer's Disease. J Alzheimers Dis 2017;57:1105-21. [Crossref] [PubMed]
- Butterfield DA. Perspectives on Oxidative Stress in Alzheimer's Disease and Predictions of Future Research Emphases. J Alzheimers Dis 2018;64:S469-79. [Crossref] [PubMed]
- Poprac P, Jomova K, Simunkova M, et al. Targeting Free Radicals in Oxidative Stress-Related Human Diseases. Trends Pharmacol Sci 2017;38:592-607. [Crossref] [PubMed]
- Vaz M, Silvestre S. Alzheimer's disease: Recent treatment strategies. Eur J Pharmacol 2020;887:173554. [Crossref] [PubMed]
- Gallardo G, Holtzman DM. Amyloid-β and Tau at the Crossroads of Alzheimer's Disease. Adv Exp Med Biol 2019;1184:187-203. [Crossref] [PubMed]
- Manoharan S, Guillemin GJ, Abiramasundari RS, et al. The Role of Reactive Oxygen Species in the Pathogenesis of Alzheimer's Disease, Parkinson's Disease, and Huntington's Disease: A Mini Review. Oxid Med Cell Longev 2016;2016:8590578. [Crossref] [PubMed]
- Awasthi S, Hindle A, Sawant NA, et al. RALBP1 in Oxidative Stress and Mitochondrial Dysfunction in Alzheimer's Disease. Cells 2021;10:3113. [Crossref] [PubMed]
- Fracassi A, Marcatti M, Zolochevska O, et al. Oxidative Damage and Antioxidant Response in Frontal Cortex of Demented and Nondemented Individuals with Alzheimer's Neuropathology. J Neurosci 2021;41:538-54. [Crossref] [PubMed]
- Chen Z, Zhong C. Oxidative stress in Alzheimer's disease. Neurosci Bull 2014;30:271-81. [Crossref] [PubMed]
- Jiang F, Zhang L, Liu Y, et al. Overexpression of miR-331 Indicates Poor Prognosis and Promotes Progression of Breast Cancer. Oncol Res Treat 2020;43:441-8. [Crossref] [PubMed]
- Angelopoulou E, Paudel YN, Piperi C. miR-124 and Parkinson's disease: A biomarker with therapeutic potential. Pharmacol Res 2019;150:104515. [Crossref] [PubMed]
- Swarbrick S, Wragg N, Ghosh S, et al. Systematic Review of miRNA as Biomarkers in Alzheimer's Disease. Mol Neurobiol 2019;56:6156-67. [Crossref] [PubMed]
- Su L, Li R, Zhang Z, et al. Identification of altered exosomal microRNAs and mRNAs in Alzheimer's disease. Ageing Res Rev 2022;73:101497. [Crossref] [PubMed]
- Chen ML, Hong CG, Yue T, et al. Inhibition of miR-331-3p and miR-9-5p ameliorates Alzheimer's disease by enhancing autophagy. Theranostics 2021;11:2395-409. [Crossref] [PubMed]
- Ma H, Huang C, Huang Q, et al. Circular RNA circ_0014717 Suppresses Hepatocellular Carcinoma Tumorigenesis Through Regulating miR-668-3p/BTG2 Axis. Front Oncol 2020;10:592884. [Crossref] [PubMed]
- Gao Z, Gao Q, Lv X. MicroRNA-668-3p Protects Against Oxygen-Glucose Deprivation in a Rat H9c2 Cardiomyocyte Model of Ischemia-Reperfusion Injury by Targeting the Stromal Cell-Derived Factor-1 (SDF-1)/CXCR4 Signaling Pathway. Med Sci Monit 2020;26:e919601. [Crossref] [PubMed]
- Guo Y, Lu G, Mao H, et al. miR-133b Suppresses Invasion and Migration of Gastric Cancer Cells via the COL1A1/TGF-β Axis. Onco Targets Ther 2020;13:7985-95. [Crossref] [PubMed]
- Ding Y, Wang L, Zhao Q, et al. MicroRNA-93 inhibits chondrocyte apoptosis and inflammation in osteoarthritis by targeting the TLR4/NF-κB signaling pathway. Int J Mol Med 2019;43:779-90. [PubMed]
- Zhang Z, Li J, Huang Y, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif 2018;51:e12505. [Crossref] [PubMed]
- Liu J, Jiang J, Hui X, et al. Mir-758-5p Suppresses Glioblastoma Proliferation, Migration and Invasion by Targeting ZBTB20. Cell Physiol Biochem 2018;48:2074-83. [Crossref] [PubMed]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351-79. [Crossref] [PubMed]
- Volkert MR, Crowley DJ. Preventing Neurodegeneration by Controlling Oxidative Stress: The Role of OXR1. Front Neurosci 2020;14:611904. [Crossref] [PubMed]
- Wang J, Rousseau J, Kim E, et al. Loss of Oxidation Resistance 1, OXR1, Is Associated with an Autosomal-Recessive Neurological Disease with Cerebellar Atrophy and Lysosomal Dysfunction. Am J Hum Genet 2019;105:1237-53. [Crossref] [PubMed]
- Dianat M, Radan M, Mard SA, et al. Contribution of reactive oxygen species via the OXR1 signaling pathway in the pathogenesis of monocrotaline-induced pulmonary arterial hypertension: The protective role of Crocin. Life Sci 2020;256:117848. [Crossref] [PubMed]
- Jiang Y, Liu J, Chen L, et al. Serum secreted miR-137-containing exosomes affects oxidative stress of neurons by regulating OXR1 in Parkinson's disease. Brain Res 2019;1722:146331. [Crossref] [PubMed]
- Shafi O. Inverse relationship between Alzheimer's disease and cancer, and other factors contributing to Alzheimer's disease: a systematic review. BMC Neurol 2016;16:236. [Crossref] [PubMed]
- Farmer KM, Ghag G, Puangmalai N, et al. P53 aggregation, interactions with tau, and impaired DNA damage response in Alzheimer's disease. Acta Neuropathol Commun 2020;8:132. [Crossref] [PubMed]
- Jazvinšćak Jembrek M, Slade N, Hof PR, et al. The interactions of p53 with tau and Aß as potential therapeutic targets for Alzheimer's disease. Prog Neurobiol 2018;168:104-27. [Crossref] [PubMed]
- Tan M, Wang S, Song J, et al. Combination of p53(ser15) and p21/p21(thr145) in peripheral blood lymphocytes as potential Alzheimer's disease biomarkers. Neurosci Lett 2012;516:226-31. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Wojsiat J, Laskowska-Kaszub K, Alquézar C, et al. Familial Alzheimer's Disease Lymphocytes Respond Differently Than Sporadic Cells to Oxidative Stress: Upregulated p53-p21 Signaling Linked with Presenilin 1 Mutants. Mol Neurobiol 2017;54:5683-98. [Crossref] [PubMed]
- Tramutola A, Pupo G, Di Domenico F, et al. Activation of p53 in Down Syndrome and in the Ts65Dn Mouse Brain is Associated with a Pro-Apoptotic Phenotype. J Alzheimers Dis 2016;52:359-71. [Crossref] [PubMed]
- Bialopiotrowicz E, Szybinska A, Kuzniewska B, et al. Highly pathogenic Alzheimer's disease presenilin 1 P117R mutation causes a specific increase in p53 and p21 protein levels and cell cycle dysregulation in human lymphocytes. J Alzheimers Dis 2012;32:397-415. [Crossref] [PubMed]
- Cui GH, Zhu J, Wang YC, et al. Effects of exosomal miRNAs in the diagnosis and treatment of Alzheimer's disease. Mech Ageing Dev 2021;200:111593. [Crossref] [PubMed]
- Walgrave H, Zhou L, De Strooper B, et al. The promise of microRNA-based therapies in Alzheimer's disease: challenges and perspectives. Mol Neurodegener 2021;16:76. [Crossref] [PubMed]
- He L, Chen Z, Wang J, et al. Expression relationship and significance of NEAT1 and miR-27a-3p in serum and cerebrospinal fluid of patients with Alzheimer's disease. BMC Neurol 2022;22:203. [Crossref] [PubMed]
- Luo G, Wang X, Liu C. MiR-483-3p improves learning and memory abilities via XPO1 in Alzheimer's disease. Brain Behav 2022;e2680. [Crossref] [PubMed]
- Mayo S, Benito-León J, Peña-Bautista C, et al. Recent Evidence in Epigenomics and Proteomics Biomarkers for Early and Minimally Invasive Diagnosis of Alzheimer's and Parkinson's Diseases. Curr Neuropharmacol 2021;19:1273-303. [Crossref] [PubMed]
- Chang WS, Wang YH, Zhu XT, et al. Genome-Wide Profiling of miRNA and mRNA Expression in Alzheimer's Disease. Med Sci Monit 2017;23:2721-31. [Crossref] [PubMed]
- Maslov LN, Naryzhnaia NV, Podoksenov IuK, et al. Reactive oxygen species are triggers and mediators of an increase in cardiac tolerance to impact of ischemia-reperfusion. Ross Fiziol Zh Im I M Sechenova 2015;101:3-24. [PubMed]
- Dai SH, Chen T, Wang YH, et al. Sirt3 protects cortical neurons against oxidative stress via regulating mitochondrial Ca2+ and mitochondrial biogenesis. Int J Mol Sci 2014;15:14591-609. [Crossref] [PubMed]
- Hambright WS, Fonseca RS, Chen L, et al. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol 2017;12:8-17. [Crossref] [PubMed]
- Zeng J, Zhu L, Liu J, et al. Metformin Protects against Oxidative Stress Injury Induced by Ischemia/Reperfusion via Regulation of the lncRNA-H19/miR-148a-3p/Rock2 Axis. Oxid Med Cell Longev 2019;2019:8768327. [Crossref] [PubMed]
- Jiang H, Niu F, Zheng Y, et al. CART mitigates oxidative stress and DNA damage in memory deficits of APP/PS1 mice via upregulating β-amyloid metabolism-associated enzymes. Mol Med Rep 2021;23:280. [Crossref] [PubMed]
- Zhang Z, Han K, Wang C, et al. Dioscin Protects against Aβ1-42 Oligomers-Induced Neurotoxicity via the Function of SIRT3 and Autophagy. Chem Pharm Bull (Tokyo) 2020;68:717-25. [Crossref] [PubMed]
- Lee KH, Lee SJ, Lee HJ, et al. Amyloid β1-42 (Aβ1-42) Induces the CDK2-Mediated Phosphorylation of Tau through the Activation of the mTORC1 Signaling Pathway While Promoting Neuronal Cell Death. Front Mol Neurosci 2017;10:229. [Crossref] [PubMed]
- Lei B, Liu J, Yao Z, et al. NF-κB-Induced Upregulation of miR-146a-5p Promoted Hippocampal Neuronal Oxidative Stress and Pyroptosis via TIGAR in a Model of Alzheimer's Disease. Front Cell Neurosci 2021;15:653881. [Crossref] [PubMed]
- Wang Q, Ge X, Zhang J, et al. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer's disease via inhibition of the miR-375/SIX4 axis. Aging (Albany NY) 2020;12:23974-95. [Crossref] [PubMed]
- Yang M, Lin X, Segers F, et al. OXR1A, a Coactivator of PRMT5 Regulating Histone Arginine Methylation. Cell Rep 2020;30:4165-4178.e7. [Crossref] [PubMed]
- Yang M, Luna L, Sørbø JG, et al. Human OXR1 maintains mitochondrial DNA integrity and counteracts hydrogen peroxide-induced oxidative stress by regulating antioxidant pathways involving p21. Free Radic Biol Med 2014;77:41-8. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


