
Association between EGFR gene mutant protein expression and T790M mutation after first-generation EGFR-TKI treatment resistance: a retrospective, single-arm clinical study
Introduction
With the development of precision medicine, gene detection is more and more widely used. Targeted therapy has become an important treatment method for patients with advanced lung cancer (1). Driver gene variation can be detected in the tumor tissue of most patients with lung adenocarcinoma, including epidermal growth factor receptor (EGFR) mutation, anaplastic lymphoma kinase (ALK) rearrangement (2), ROS-1 rearrangement (3), rearranged during transfection (RET) rearrangement, mesenchymal-epithelial transition (MET) exon 14 deletion (4), KRAS mutation, and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α (PIK3CA) mutation, among others. Targeted therapy based on these driver genes has greatly improved therapeutic efficacy in patients with advanced lung adenocarcinoma (5).
EGFR is a receptor tyrosine kinase that is widely distributed on the surface of mammalian epithelial cells and is involved in cell proliferation and differentiation. EGFR binds to its ligand to form a dimer. It regulates the growth and reproduction of tumor cells by activating downstream signaling pathways through autophosphorylation, including the mitogen-activated protein kinase (MAPK) pathway and phosphatidylinositol 3-kinase (PI3K)/AKT pathway, among others (6). Multiple studies have shown that EGFR mutations are common in Asian, nonsmoker or mild smoker, female adenocarcinoma patients who may benefit from EGFR-tyrosine kinase inhibitor (TKI)-targeted therapy. The 2009 Iressa Pan-Asia Study (IPASS) compared gefitinib with carboplatin/paclitaxel in patients with advanced non-small cell lung cancer. It showed that gefitinib had significantly longer progress-free survival than carboplatin/paclitaxel and confirmed that EGFR gene mutations could be used as biomarkers to evaluate gefitinib efficacy (7). EGFR-TKI is approved for advanced non-small cell lung cancer with EGFR mutations.
EGFR-TKIs are an important treatment option for patients with EGFR-mutated lung adenocarcinoma. However, most patients eventually develop acquired resistance after 1 year, including T790M mutation (8), MET amplification, human epidermal growth factor receptor 2 (HER-2) amplification (9), and PIK3CA mutation (10). T790m mutation is currently known to be a common cause of acquired drug resistance following first-generation TKIs and an effective target for third-generation EGFR-TKIs. However, it is unknown which patients will develop the T790M mutation following first-generation TKIs. The IPASS study systematically compared the predictive value of EGFR gene mutations and mutant protein expression for the response to first-generation EGFR-TKI treatment. However, it failed to observe the association between the T790M mutation and the above markers after first-generation EGFR-TKI resistance. Although third-generation EGFR-TKIs are becoming the standard of care for EGFR mutation-positive NSCLC patients worldwide, first- or second-generation EGFR-TKIs followed by third-generation TKIs may offer longer survival, so predicting T790M positivity is highly suggestive. In this study, we used immunohistochemistry (IHC) to detect the expression of EGFR protein in tumor tissues of patients with stage IV lung adenocarcinoma who had a known EGFR mutation, analyzed the relationship between protein expression and the efficacy of the first-generation TKI, and investigated the incidence of T790M mutation in patients who developed drug resistance. The aim of this research was to explore the population characteristics of patients with T790M mutation and to try to find markers that could be used to predict T790M resistance after first-generation EGFR-TKI treatment. The findings are summarized below. We present the following article in accordance with the REMARK reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3850/rc).
Methods
Study subjects
The tumor tissue samples and clinical treatment data of stage IV lung adenocarcinoma patients who visited The First Affiliated Hospital of Anhui Medical University and The Second Affiliated Hospital of Anhui Medical University between January 1, 2018 and March 30, 2021 were analyzed retrospectively in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Commission of the First Affiliated Hospital of Anhui Medical University (PJ2022-10-43). Written informed consent to participate in this study was provided by all participants/next of kin.
The criteria for inclusion were: aged ≥18 years old; pathologically confirmed adenocarcinoma; stage IV patients with distant organ metastasis at the time of oral EGFR-TKI; tumor tissue samples confirmed by sequencing to have EGFR mutations, including exon 19 deletion and L858R point mutation; received first-generation EGFR-TKI targeted therapy for at least 2 months, including gefitinib, icotinib, and erlotinib; with RECIST 1.1-compliant target lesions; and Eastern Cooperative Oncology Group (ECOG) score 0–1.
The exclusion criteria were: second and third-generation EGFR-TKIs after first-line EGFR-TKI therapy; patients with other malignancies; patients with EGFR non-19 exon deletion or L858R point mutation; and patients whose efficacy could not be determined (for example, due to no measurable lesions, lack of imaging data, and loss to follow-up).
Immunohistochemical assays
The Department of Pathology reviewed all tumor tissue samples, and the proportion of tumor cells met the quality control criteria. After quality control, the content of tumor cells in the submitted samples shall not be lower than the detection standard (detection standard: proportion of tumor cells ≥20%, number of tumor cells ≥100). Protein antibody against EGFR 19 exon deletion (E746-A750del) and EGFR 21 exon mutation (L858R) were rabbit monoclonal antibodies (clone numbers SP111, SP125; Fuzhou Maixin Biotechnology Development Co., Ltd., Beijing, China), and secondary antibodies were enzyme-labeled goat anti-mouse/rabbit immunoglobulin G (IgG) polymers (Fuzhou Maixin Biotechnology Development Co., Ltd.). The mouse/rabbit MaxVision-HRP kit was used. Paraffin sections (4 µm) were cut, routinely deparaffinized, hydrated, and repaired at high temperature using ethylenediaminetetraacetic acid (EDTA) antigen retrieval solution. Blocking agent was added to block endogenous peroxidase, and primary antibody reagent was incubated overnight at 4 ℃. The samples were rinsed, incubated with secondary antibody reagent at 37 ℃ for 15 minutes, developed by 3,3’-diaminobenzidine (DAB), counterstained with hematoxylin, rinsed, and dehydrated with gradient ethanol.
Evaluation of immunohistochemical results
The protein expressions were evaluated after the NSCLC was diagnosed. The immunohistochemical results were judged independently by two pathologists. The presence of yellow or brownish-yellow staining in the membrane/cytoplasm of the tumor tissue without background staining was considered positive, and the absence of any yellow or brownish-yellow staining in the tumor tissue was considered negative.
Evaluation of efficacy
Patients who met all of the inclusion and exclusion criteria had their medical histories collected retrospectively, including information on the use of first-generation TKIs, progression-free survival (PFS) for first-generation TKIs, and T790M mutation detection results in peripheral blood by digital polymerase chain reaction (PCR) or Super-ARMS after first-generation TKI resistance. The time from the first dose to radiographic progression was defined as PFS. The progression refers to the minimum sum of diameters of all target lesions measured in the whole treatment process as a reference, and the relative increase of diameters and diameters should be at least 20%, and the absolute increase of diameters and diameters should be at least 5 mm. If one or more new lesions appear, the disease progression is also considered.
Statistical analysis
The statistical software packages SPSS for Windows, software version 25.0 (SPSS Inc., Chicago, IL, USA) and Graphpad Prism 8.0 (GraphPad Software, San Diego, CA, USA) were used for analysis. Enumeration data were statistically described by relative number (percentage), and the χ2 test was used for comparison. The Kaplan-Meier method was used to calculate PFS curves, and differences were compared using the log-rank test, with P<0.05 indicating a statistically significant difference.
Role of the funding source
The funding agencies did not participate in study design, data collection, data analysis, or manuscript writing. The corresponding authors were responsible for all aspects of the study to ensure that issues related to the accuracy or integrity of any part of the work were properly investigated and resolved. The final version was approved by all authors.
Results
Basic characteristics of the study population
This study included 69 stage IV lung adenocarcinoma patients (aged 40–81 years, median age 67 years) who met the inclusion criteria, had complete medical history data, and had peripheral blood T790M detection after drug resistance (Table 1).
Table 1
| Clinical data | 19 exon deletion | L858R point mutation | χ2 value | P value |
|---|---|---|---|---|
| Gender | 2.077 | 0.150 | ||
| Male | 12 | 10 | ||
| Female | 17 | 30 | ||
| Age (years) | 0.960 | 0.327 | ||
| ≤65 | 12 | 12 | ||
| >65 | 17 | 28 | ||
| Medicine | ||||
| Gefitinib | 13 | 24 | 1.556 | 0.212 |
| Icotinib | 12 | 11 | 1.457 | 0.227 |
| Erlotinib | 4 | 5 | 0.025 | 0.875 |
| Protein expression of 19del/L858R when diagnosed as NSCLC | 7.035 | 0.008 | ||
| Positive | 11 | 28 | ||
| Negative | 18 | 12 | ||
| T790M expression after TKI resistance | 0.146 | 0.703 | ||
| Yes | 11 | 17 | ||
| No | 18 | 23 | ||
NSCLC, non-small cell lung cancer; TKI, tyrosine kinase inhibitor.
Protein expression
There were no statistically significant differences in efficacy between the groups with positive and negative protein expression. The median PFS (mPFS) of all patients with exon 19 deletion was 13.20 months (95% CI: 11.18–15.23 months), including 13.20 months (95% CI: 10.45–15.95 months) in the protein expression-positive group and 13.00 months (95% CI: 2.67–23.33 months) in the protein expression-negative group, with no statistically significant difference, χ2=0.405, P=0.817 (Figure 1). The mPFS of all patients with L858R point mutation was 12.13 months (95% CI: 8.10–16.16 months), including 12.13 months (95% CI: 7.50–16.76 months) in the protein expression-positive group and 11.93 months (95% CI: 9.48–14.39 months) in the protein expression-negative group, with no statistically significant difference, χ2=0.191, P=0.909 (Figure 2).
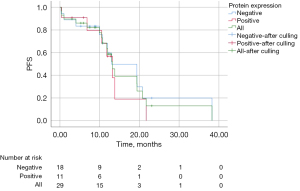
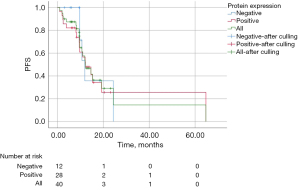
T790M mutation and protein expression
We analyzed the T790M mutation in peripheral blood after resistance to first-generation EGFR-TKI treatment, and the results showed that the protein expression-positive group was more likely to produce the T790M mutation after resistance, and the difference was statistically significant. In patients with exon 19 deletion, T790M mutation developed in 7 (63.64%, 7/11) patients in the positive protein expression group and 4 (22.22%, 4/18) in the negative protein expression group (χ2=4.974, P=0.026). In patients with L858R point mutation, there were 15 (53.57%, 15/28) and 2 (16.67%, 2/12) in the positive and negative protein expression groups, respectively (χ2=4.682, P=0.030) (Figure 3).
Discussion
Lung cancer is a deadly malignant tumor that affects millions of people worldwide. A common type of driver gene mutation in lung adenocarcinoma is the EGFR gene mutation. Exon 19 deletion and exon 21 L858R point mutation are 2 common EGFR effective mutations (11). The IPASS study published in 2009 found that gefitinib treatment for patients with EGFR gene mutations resulted in significantly longer PFS time than chemotherapy (7).
EGFR gene mutation, gene copy number, and protein expression are all common biomarkers used to predict the efficacy of EGFR-TKIs, and they frequently overlap (12). The relationship between the 3 biomarkers and the efficacy of targeted therapy was investigated in the IPASS study’s biomarker analysis. The results showed that in patients with a positive EGFR gene mutation, gefitinib significantly prolonged PFS and increased overall response rate (ORR) compared to chemotherapy. In the protein expression analysis, the gefitinib group had PFS in patients with positive protein expression, but there was no significant difference in ORR and overall survival (OS) between the two groups (13). As a result, EGFR mutations distinguished the benefit population more effectively than EGFR protein expression, establishing EGFR mutations as predictive markers of response to TKI therapy.
This study re-examined the relationship between protein expression and the efficacy of targeted therapy in stage IV lung adenocarcinoma patients with known EGFR gene mutations. Among 69 patients with EGFR mutations, 39 had corresponding protein expression detected. The mPFS was similar in the protein expression-positive and negative groups, showing no statistically significant difference. This was consistent with the findings of the IPASS study, which found that protein expression had little predictive value for the efficacy of targeted therapy. In a retrospective study of patients with advanced non-small cell lung cancer (NSCLC) treated with EGFR-TKI, patients with exon 19 deletion had better clinical benefit than those with exon 21 mutation, with PFS (13.2 vs. 10.8 months, χ2=4.700, P=0.030) and OS of 30.2 and 25.6 months, respectively, in the two groups (14). It has been proposed that this difference may be related to structural changes caused by different mutations and subsequent T790M mutations (15). In this study, patients with exon 19 deletion had a slightly longer mPFS than patients with the L858R point mutation (13.20 vs. 12.13 months).
The mPFS with targeted first-generation EGFR-TKIs is now estimated to be 10–14 months (16). There are diverse mechanisms of subsequently acquired resistance, including T790M mutation, c-MET amplification (17), HER-2 amplification (18), and PIK3CA mutation (19), with the most common being the T790M mutation, which accounts for more than half of acquired resistance (20). In the phase III AURA3 study, PFS was significantly longer in the osimertinib group than in the pemetrexed/platinum group (10.10 vs. 4.40 months, hazard ratio 0.30, 95% CI: 0.23–0.41, P<0.001). In addition, the ORR was also significantly better than in the chemotherapy group (71.00% vs. 31.00%, odds ratio 5.39, 95% CI: 3.47–8.48, P<0.001) (21), suggesting that osimertinib was more effective than platinum-based combination chemotherapy for patients with NSCLC who have a T790M mutation and have progressed after first-generation EGFR-TKI therapy. For patients with NSCLC who have the T790M resistance mutation, third-generation TKIs are currently recommended (22).
Furthermore, the OS time for patients with the T790M mutation treated with first-generation or second-generation EGFR-TKIs followed by third-generation TKIs can be up to 40 months. The treatment time with EGFR-TKIs was 27.6 months (90% CI: 25.9–31.3 months) in a real-world study (NCT03370770) of afatinib and osimertinib in the sequential treatment of patients with EGFR-mutated lung cancer, and it is worth noting that the treatment time was 30.3 months for patients with exon 19 deletions and 46.7 months for Asian patients (23). This study investigated protein expression and T790M mutations in the peripheral blood of patients with subsequent drug resistance and discovered that patients with EGFR mutations and protein expression were more likely to have T790M mutations. A greater proportion of T790M mutations occurred in the positive protein expression group compared to the negative protein expression group (63.64% vs. 22.22%, Figure 2; χ2=4.974, P=0.026). In patients with L858R point mutations, the T790M mutation rates were 53.57% and 16.67% in the positive and negative protein expression groups, respectively (χ2=4.682, P=0.030). Although the IPASS study found that protein expression had little predictive value for the efficacy of targeted therapy, the T790M mutation was not investigated. Suppose patients with an EGFR gene mutation and protein expression are more likely to have a T790M mutation. In that case, after patients became resistant to first-generation or second-generation EGFR-TKI, patients with T790M mutation showed significantly better efficacy with third-generation EGFR-TKI than those without T790M mutation. The patients with EGFR gene mutations and protein expression were more likely to have T790M mutations after first-generation EGFR-TKI treatment progression. As a result, EGFR protein expression could be used to predict the efficacy of third-generation TKIs and select the candidate to receive afatinib-osimertinib sequential therapy. However, because this was a retrospective study with a small sample size and potentially skewed results, additional prospective and multicenter studies are required for validation.
Conclusions
The efficacy of EGFR-TKI as first-line treatment for lung adenocarcinoma with EGFR gene mutation is not related to protein expression, similar to the results from IPASS study. However, this study discovered that patients with EGFR gene mutations and protein expression were more likely to have T790M mutations after first-generation EGFR-TKI treatment progression, and these patients benefited from third-generation TKI treatment.
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group.
Funding: This work was supported by the Beijing SisKE Clinical Oncology Research Foundation (No. Y-Q201802-047).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3850/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3850/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3850/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Commission of the First Affiliated Hospital of Anhui Medical University (PJ2022-10-43). Written informed consent to participate in this study was provided by all participants/next of kin.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-59. [Crossref] [PubMed]
- Heriyanto DS, Trisnawati I, Kumara EG, et al. The Prevalence of the EML4-ALK Fusion Gene in Cytology Specimens from Patients with Lung Adenocarcinoma. Pulm Med 2020;2020:3578748. [Crossref] [PubMed]
- Shaw AT, Riely GJ, Bang YJ, et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): updated results, including overall survival, from PROFILE 1001. Ann Oncol 2019;30:1121-6. [Crossref] [PubMed]
- Wang Y, Tian P, Xia L, et al. The clinical efficacy of combinatorial therapy of EGFR-TKI and crizotinib in overcoming MET amplification-mediated resistance from prior EGFR-TKI therapy. Lung Cancer 2020;146:165-73. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Dong RF, Zhu ML, Liu MM, et al. EGFR mutation mediates resistance to EGFR tyrosine kinase inhibitors in NSCLC: From molecular mechanisms to clinical research. Pharmacol Res 2021;167:105583. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Suda K, Onozato R, Yatabe Y, et al. EGFR T790M mutation: a double role in lung cancer cell survival? J Thorac Oncol 2009;4:1-4. [Crossref] [PubMed]
- Cretella D, Saccani F, Quaini F, et al. Trastuzumab emtansine is active on HER-2 overexpressing NSCLC cell lines and overcomes gefitinib resistance. Mol Cancer 2014;13:143. [Crossref] [PubMed]
- Qu GP, Shi M, Wang D, et al. Dual targeting of MEK and PI3K effectively controls the proliferation of human EGFR-TKI resistant non-small cell lung carcinoma cell lines with different genetic backgrounds. BMC Pulm Med 2021;21:208. [Crossref] [PubMed]
- Zhang Y, He D, Fang W, et al. The Difference of Clinical Characteristics Between Patients With Exon 19 Deletion and Those With L858R Mutation in Nonsmall Cell Lung Cancer. Medicine (Baltimore) 2015;94:e1949. [Crossref] [PubMed]
- Uramoto H, Mitsudomi T. Which biomarker predicts benefit from EGFR-TKI treatment for patients with lung cancer? Br J Cancer 2007;96:857-63. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Jiang H, Zhu M, Li Y, et al. Association between EGFR exon 19 or exon 21 mutations and survival rates after first-line EGFR-TKI treatment in patients with non-small cell lung cancer. Mol Clin Oncol 2019;11:301-8. [Crossref] [PubMed]
- Zhang Y, Sheng J, Kang S, et al. Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: a meta-analysis. PLoS One 2014;9:e107161. [Crossref] [PubMed]
- Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [Crossref] [PubMed]
- Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009;20:298-304. [Crossref] [PubMed]
- Liu S, Li S, Hai J, et al. Targeting HER2 Aberrations in Non-Small Cell Lung Cancer with Osimertinib. Clin Cancer Res 2018;24:2594-604. [Crossref] [PubMed]
- Ludovini V, Bianconi F, Pistola L, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:707-15. [Crossref] [PubMed]
- Wu SG, Liu YN, Tsai MF, et al. The mechanism of acquired resistance to irreversible EGFR tyrosine kinase inhibitor-afatinib in lung adenocarcinoma patients. Oncotarget 2016;7:12404-13. [Crossref] [PubMed]
- Mok TS, Wu Y-L, Ahn M-J, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Yi L, Fan J, Qian R, et al. Efficacy and safety of osimertinib in treating EGFR-mutated advanced NSCLC: A meta-analysis. Int J Cancer 2019;145:284-94. [Crossref] [PubMed]
- Hochmair MJ, Morabito A, Hao D, et al. Sequential treatment with afatinib and osimertinib in patients with EGFR mutation-positive non-small-cell lung cancer: an observational study. Future Oncol 2018;14:2861-74. [Crossref] [PubMed]
(English Language Editor: A. Muylwyk)



