
Rutaecarpine prevents the malignant biological properties of breast cancer cells by the miR-149-3p/S100A4 axis
Introduction
Breast cancer (BC) is one of the most common cancers in women, and accounts for 30% of all female cancers (1). BC ranks 1st in morbidity and 2nd in mortality among female malignancies (2). A growing number of studies have shown that the main causes of BC include artificial abortion, the use of clomiphene citrate and gonadotropin, ionizing radiation, alcohol abuse, and smoking (3). Currently, the therapy modalities of BC mainly include surgery, chemotherapy, radiotherapy, bio-targeted therapy, hormone therapy, and immunotherapy (4). As the prevention and treatment methods for BC continue to improve, the mortality of BC patients continues to decline. However, some patients suffer from recurrence and metastasis for which the therapeutic effect is still poor. BC is a complex polygenic disease (5). Thus, in-depth explorations of the risk factors and internal pathogenesis of BC will have far-reaching effects in the prevention and therapy of BC.
Rutaecarpine (RUT), which is an indole quinazoline alkaloid, is one of the main active components in evodiae fructus (6). Recent research has confirmed that RUT exerts anti-tumor effects through multiple pathways, promoting tumor cell apoptosis and cell-cycle arrest and inhibiting tumor cell neovascularization (7). However, the effects and mechanism of RUT in the human BC process are still unclear. The further elucidation of the mechanism of RUT in BC is critical for the treatment of BC.
Micro ribonucleic acid (miRNA) is an endogenous, non-coding RNA, about 22 nt in length (8). MiRNA silences or degrades messenger RNA (mRNA) translation by binding to the target mRNA 3 prime untranslated region (9). Numerous miRNAs have been shown to play therapeutic roles in advanced cancers, including miR-124 in BC (10), miR-29 and miR-200a in ovarian cancer (11), and miR-21 in cervical cancer (12). In our pre-experiments, we found that miR-149-3p high expression was associated with long patient survival time by bioinformatics, and miR-149-3p was downregulated in BC cells by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) verification. MiR-149-3p, a miRNA, has been shown to have an obvious inhibitory role on the progression of various cancers, including bladder cancer (13), lung cancer (14), and BC (15). However, the role and mechanism of miR-149-3p in the apoptosis and autophagy of BC cells are not clear, which is also the aim of our current study.
It has been reported that RUT regulates numerous downstream genes, such as vascular endothelial growth factor receptor 2 (VEGFR2) (16), Notch1 (17), NADPH oxidase 4 (NOX4) (18), A disintegrin and metalloproteinase 17 (ADAM17) (18), Transient receptor potential vanilloid member 1 (TRPV1) (19), calcitonin-gene related peptide (CGRP) (20), and adenosine monophosphate-activated protein kinase (AMPK) (21). Studies have also shown that Notch1 interacts with protein 53 (p53) (22), and p53 binds to the Nox4 promoter (23). P53, which is a key tumor suppressor gene, indirectly regulates miRNA. For example, wild-type (WT) p53 positively regulates miR-149-3p (24). Thus, we speculated that RUT would upregulate miR-149-3p. However, it was not clear whether RUT would block BC progression by regulating miR-149-3p.
This study sought to further investigate the effects of RUT and miR-149-3p on the biological function of BC cells and their possible mechanisms. We first screened the optimal concentration and time of RUT in BC cells and examined the expression changes of miR-149-3p in BC cells. We also examined the regulatory role of RUT in miR-149-3p in BC cells and the effects of RUT and miR-149-3p on the proliferation, cycle, apoptosis, autophagy, and angiogenesis of BC cells. Further, we explored the possible target genes and regulatory pathways of miR-149-3p in BC. Our findings may provide effective drugs and targets for BC therapy. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3765/rc).
Methods
Tissue samples
Sentinel lymph node and BC tissues were collected from 5 pairs of BC patients, who were diagnosed at the Wuhan Asia General Hospital between 2019 and 2020. None of the patients had undergone surgery, radiation, or chemotherapy. After collection, all the tissues were preserved in liquid nitrogen. All the patients who participated in this study signed an informed consent form, and this study was approved by the Institutional Ethics Board of Wuhan Asia General Hospital (No. WAGHMEC-KY-2022009). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
human normal breast cells (MCF-10A; Cat. No. CRL-10317), and four breast cancer cell lines (ZR-75-1, Cat. No. CRL-1500; BCAP-37, Cat. No. CL1026; MCF-7, Cat. No. CL1123; and MDA-MB-231, Cat. No. HTB-26) were all provided by ATCC. MCF-10A and MCF-7 cells were cultured in Dulbecco’s modified eagle medium (DMEM; Gibco, Cat. No. 12800017) with 10% fetal bovine serum (FBS, Sigma, Welwyn Garden City, UK) at 37 ℃ with 5% carbon dioxide (CO2). ZR-75-1, BCAP-37, and MDA-MB-231 cells were incubated in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Cat. No. 31800022), including 10% FBS at 37 ℃ with 5% CO2.
Cell transfection and treatment
The negative control (NC), miR-149-3p inhibitors, and miR-149-3p mimics were provided by GenePharma (Suzhou, China). S100A4 complementary deoxyribonucleic acid (cDNA) was generated through polymerase chain reaction (PCR) using the following primers: S100A4-XhoI, 5'-ccgctcgagAAACTCCTCTGATGTGGTTGGGGGGTCTGCCAG-3', and S100A4-NotRI, 5'-ataagaatgcggccgcAAAACTTCCAAGAATCTTTATTGAACTTG-3'. Then the amplified fragment (138 bp) was cloned into the psiCHECK-2 plasmid. The MDA-MB-231 cells (1×105 cells/well) were evenly distributed in a 6-well plate for 8 h. Subsequently, the cells were transfected with the NC, miR-149-3p inhibitor, or S100A4-overexpressed plasmid with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for 48 h in accordance with the manufacturer’s instructions. Next, the transfected cells were processed with the autophagy agonist (rapamycin, Solarbio, Cat. No. R8140-25) or the angiogenesis inhibitor (TNP-470, TargetMol, USA, Cat. No. T17110). The BC cells were treated with 0, 1, 5, 10, 20, and 40 µM RUT for 0, 12, 24, 48, and 72 h (25,26).
RT-qPCR
The BC cells in each group were collected, and Trizol (Invitrogen, MA, USA) was added to acquire the total RNAs using the conventional method. Next, we synthesized the cDNA using a reverse transcription kit (Takara, Tokyo, Japan), and the procedure was conducted in accordance with the kit instructions. qPCR was carried out using the SYBR Green qPCR Master Mix (DBI Bioscience, cat. no. DBI-2043). The results were calculated using the 2−△△CT method. The primer sequences are listed in Table 1.
Table 1
| ID | Sequence (5'-3') |
|---|---|
| GAPDH (forward) | TGTTCGTCATGGGTGTGAAC |
| GAPDH (reversed) | ATGGCATGGACTGTGGTCAT |
| S100A4 (forward) | GAAGACTCTCTTGTCTGTCGGA |
| S100A4 (reversed) | AATGGCGGTACTGACTTGATG |
| U6 (forward) | CTCGCTTCGGCAGCACA |
| U6 (reversed) | AACGCTTCACGAATTTGCGT |
| MiR-149-3p (forward) | CGGGCGAGGGAGGGACGGGG |
| MiR-149-3p (reversed) | CAGCCACAAAAGAGCACAAT |
RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Western blot
After cleaning, the treated cells were cleaved by adding radio-immunoprecipitation assay (RIPA) (Beyotime, China) on ice. After centrifugation at 4 ℃, the supernatant solution was obtained. The protein samples were quantitatively tested using the bicinchoninic acid (BCA) method. After being boiled at 100 ℃ for 5 min with buffer, the protein was used for electrophoresis with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membrane (Millipore, Cat. No. IPVH00010). After blocking, the membranes were processed with the primary antibody overnight at 4 ℃ and the secondary antibody (1:10,000, Southern Biotech, China) for 1 h. The membranes were exposed to electrochemiluminescence (ECL) reagent (Millipore, Cat. No. WBKLS0500), and the blots were developed using X-ray film (Kodak, Rochester, NY, Cat. No. XBT-1).
CCK-8
The MDA-MB-231 cells were collected and counted with a cell counting plate. After treatment, the BC cells (5×103 cells/well, 100 µL/well) were inoculated in 96-well plate and cultured at 37 ℃ with 5% CO2. Next, 10 µL of Cell Counting Kit-8 (CCK-8) reagent (Dojindo, Japan) was added to each well at the specified time point. After the 96-well plate was further incubated for 3 h, cell absorbance was determined using a microplate reader.
Clone formation assays
The MDA-MB-231 cells in each group were evenly added to a 6-well plate with 500 cells/well. All the cells were routinely cultured for 14 days at 37 ℃. After washing, the cells in each group were fixed and stained using 0.2% crystal violet. After washing and drying, the cell clones were observed and photographed under a light microscope.
Flow cytometry
The treated BC cells were harvested and suspended with phosphate buffered solution. The MDA-MB-231 cells were fixed using 500 µL of 70% ethyl alcohol. After centrifugation, the cells were suspended and incubated with 100 µL of RNase at 37 ℃ for 30 min and dyed with 200 µL of V propidium iodide (PI) for 20 min. The cell-cycle rates were determined by flow cytometry (BD Biosciences, USA).
Luciferase reporter assays
On account of the predicted sites between miR-149-3p and S100A4, we constructed the WT and mutant (Mut) S100A4 plasmids with the psiCHECK-2 vector. The primer sequences for Mut-S100A4 were 5'-GTCTGCCAGCTGGGGTTTGTTTTGTCGCCAGTGGGCACTTTTTTTTTTCCACCCTG-3' (forward), 5'-GTGCCCACTGGCGACAAAACAAACCCCAGCTGGCAGACCCCCCAACCACATCAGAG-3' (reverse). Subsequently, the MDA-MB-231 cells were co-transfected with miR-149-3p mimics or a miR-149-3p inhibitor and WT-S100A4 or Mut-S100A4 using Lipofectamine 3000 (Invitrogen) for 48 h. Subsequently, the dual-luciferase assay kit (Promega, Madison, WI, USA) was used to evaluate the luciferase activity in accordance with the manufacturer’s instructions.
Immunohistochemistry (IHC) assays
As described previously (27), the paraffin-embedded tissue was cut into 4-µm thick sections, which were then immersed in EDTA solution at pH8.0 for 10 min before undergoing microwave antigen repair. After washing, goat serum was successively added to the sections for 20 min. Then the sections were treated with anti-S100A4 (Abcam, Cambridge, UK; ab58597) overnight at 4 ℃ and secondary antibody (Abcam) for 30 min. A diffuse alveolar damage (DAB) kit was applied to treat the sections. Then the tissues were examined using an inverted microscope after neutral gum sealing.
Statistical analysis
The data are represented as the mean ± standard deviation. The statistical analysis was conducted with SPSS21. 0 (SPSS, Inc., Chicago, IL, USA), and a 1-way analysis of variance was also performed. A P value <0.05 indicated a statistically significant difference in the results.
Results
The expression levels of miR-149-3p and S100A4 in BC cells
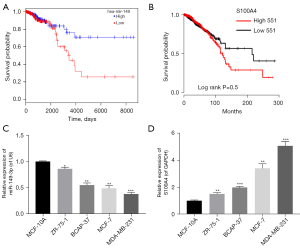
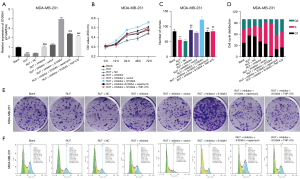
In line with The Cancer Genome Atlas (TCGA) (28), we found that the survival time in miR-149-3p high expression group was longer than that in miR-149-3p low expression group (see Figure 1A). Conversely, the high expression of S100A4 was correlated with a short survival time, and thus indicated a poor prognosis (see Figure 1B). Somewhat unexpectedly, we discovered that miR-149-3p was significantly downregulated in BC cells, including the ZR-75-1, BCAP-37, MCF-7, and MDA-MB-231 cells, and most especially, the MDA-MB-231 cells (see Figure 1C). Additionally, the RT-qPCR results revealed that relative to the MCF-10A cells, S100A4 was significantly upregulated in the 4 BC cell lines, especially in the MDA-MB-231 cells (see Figure 1D). Thus, we used the MDA-MB-231 cells for further experiments in this study.
Screening of the optimal concentration and time of RUT in BC cells
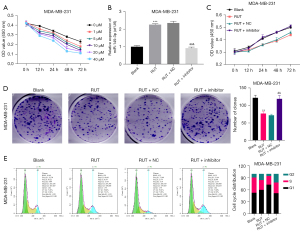
To determine whether RUT affected BC progression, the MDA-MB-231 cells were exposed to 0, 1, 5, 10, 20 and 40 µM of RUT for 0, 12, 24, 48, and 72 h. After exposure to RUT, the growth abilities of the BC cells decreased in a dose- and time-dependent manner (see Figure 2A). Based on these results, we treated the BC cells with 20 µM of RUT for 48 h in our subsequent experiments.
Inhibition of miR-149-3p significantly attenuated the inhibitory effect of RUT on the proliferation and induction of cell-cycle arrest in BC cells
We also found that RUT upregulated miR-149-3p in the MDA-MB-231 cells, while the reduction of miR-149-3p significantly weakened the upregulation of miR-149-3p mediated by RUT (see Figure 2B). Next, the CCK-8 results revealed that RUT significantly decreased cell viability, while a decrease of miR-149-3p significantly enhanced cell viability mediated by RUT in BC cells (see Figure 2C). The clone formation results indicated that RUT significantly reduced the clonality of MDA-MB-231 cells, while RUT-mediated reduction in cell clonality was significantly reversed by the miR-149-3p inhibitor (see Figure 2D). The flow cytometry results showed that RUT increased the G1-phase cells and decreased S-phase cells, while the changes in the number of G1- and S- phase cells mediated by RUT were reversed by using the miR-149-3p inhibitor in the MDA-MB-231 cells (see Figure 2E). Overall, these results showed that RUT significantly prevented proliferation and facilitated cell-cycle arrest by upregulating miR-149-3p in the MDA-MB-231 cells.
Inhibition of miR-149-3p significantly reversed the RUT-mediated expression of proteins related to apoptosis, autophagy, and angiogenesis in BC cells
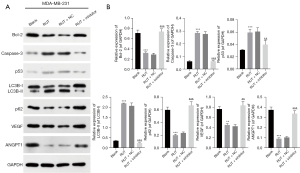
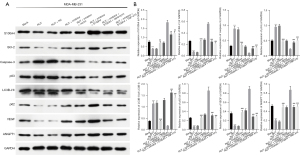
Subsequently, we further examined the effects of RUT-mediated miR-149-3p on apoptosis, autophagy, and angiogenesis-related proteins in the MDA-MB-231 cells. As Figure 3 shows, the RUT treatment significantly reduced B cell lymphoma 2 (Bcl-2), p62, vascular endothelial growth factor (VEGF), and polyclonal antibody to angiopoietin 1 (ANGPT1) expression levels, and elevated caspase-3, p53, and LC3B-II/I expression levels, while changes in the expression of these proteins were reversed by the miR-149-3p inhibitor in the MDA-MB-231 cells. Generally, these findings showed that RUT resulted in the prominent promotion of apoptosis and autophagy, and suppressed angiogenesis in the MDA-MB-231 cells.
MiR-149-3p directly targeted S100A4
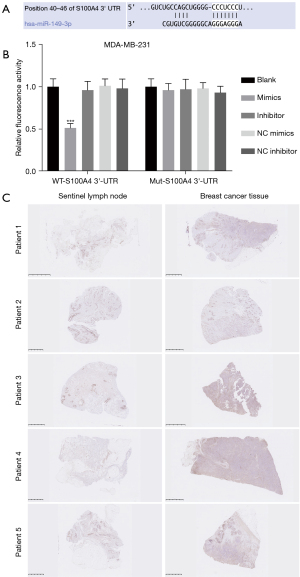
We further analyzed the possible target genes using bioinformatics software. Through screening, we discovered that there were underlying binding sites between miR-149-3p and S100A4, which has been reported as an oncogene (see Figure 4A). The luciferase reporter results showed that miR-149-3p ignorantly restrained the luciferase activity of WT-S100A4, but had no effect on that of Mut-S100A4 in the MDA-MB-231 cells (see Figure 4B). Similarly, the IHC results showed that S100A4 expression was significantly more increased in the BC tissues than the sentinel lymph node tissues (see Figure 4C). Thus, our current results showed that S100A4, as a target gene of miR-149-3p, was highly expressed in the MDA-MB-231 cells.
RUT inhibited the proliferation and accelerated cell-cycle arrest by miR-149-3p/S100A4 axis to regulate autophagy and angiogenesis pathways in BC cells
Based on the above research results, RUT upregulated miR-149-3p, S100A4 was a target gene of miR-149-3p, and RUT induced apoptosis and autophagy, and suppressed angiogenesis. We further investigated the regulatory relationships between these genes and pathways in MDA-MB-231 cells through rescue experiments. After transfection with the miR-149-3p inhibitor, the RUT-treated MDA-MB-231 cells were processed with autophagy agonists (rapamycin) or an angiogenesis inhibitor (TNP-470). We confirmed that miR-149-3p/S100A4 axis participated in the RUT-mediated blocking of BC progression by regulating autophagy and angiogenesis pathways (Figure 5). As Figure 5A shows, the reduction in miR-149-3p significantly increased S100A4 expression, which was inhibited by RUT in the MDA-MB-231 cells; the overexpression of S100A4 also significantly increased S100A4 expression, which was upregulated by the miR-149-3p inhibitor; further, the upregulation of S100A4 as mediated by S100A4 overexpression and the miR-149-3p inhibitor was also significantly attenuated by rapamycin or TNP-470 in the RUT-treated MDA-MB-231 cells. The CCK-8 results showed that suppression of miR-149-3p significantly aggrandized cell viability, which was prevented by RUT in the MDA-MB-231 cells; the overexpression of S100A4 significantly enhanced cell viability, which was induced by the miR-149-3p inhibitor; additionally, the enhancement of cell viability mediated by S100A4 overexpression and the miR-149-3p inhibitor was also reversed by rapamycin or TNP-470 in the RUT-treated MDA-MB-231 cells (see Figure 5B). Similarly, the clone formation results also showed that RUT reduced cell clonality by the miR-149-3p/S100A4/autophagy and angiogenesis pathways in the MDA-MB-231 cells (see Figure 5C,5E). Further, the flow cytometry results also showed that the overexpression of S100A4 further enhanced the decrease of G1 phase cells mediated by the miR-149-3p inhibitor, while rapamycin or TNP-470 reversed the decrease of G1 phase cells by S100A4 overexpression and the miR-149-3p inhibitor in the RUT-treated MDA-MB-231 cells (see Figure 5D,5F).
RUT regulated apoptosis, autophagy, and angiogenesis-related proteins by targeting the miR-149-3p/S100A4 axis in BC cells
We also discovered that decreased miR-149-3p increased S100A4, Bcl-2, p62, VEGF, and ANGPT1 expression levels, and decreased caspase-3, p53, and LC3B-II/I expression levels, which were mediated by RUT in the MDA-MB-231 cells. The overexpression of S100A4 further enhanced the changes in these proteins mediated by the miR-149-3p inhibitor, and rapamycin or TNP-470 also significantly reduced S100A4, Bcl-2, p62, VEGF, and ANGPT1 expression levels and elevated caspase-3, p53, and LC3B-II/I expression levels, which were mediated by S100A4 overexpression and the miR-149-3p inhibitor in the RUT-treated MDA-MB-231 cells (see Figure 6). Thus, we found that the miR-149-3p/S100A4 axis was also involved in the RUT-mediated acceleration of apoptosis and autophagy, and the inhibition of angiogenesis in the MDA-MB-231 cells.
Discussion
BC, which is a unique solid tumor, has multiple complex mechanisms. Due to the heterogeneity of BC, it cannot be detected or treated simply by conventional indicators (1). Further, most anti-cancer drugs tend to be toxic to normal breast cells or the cancer cells become resistant to the drugs (29). Traditional Chinese medicine (TCM) therapy mainly emphasizes the goal of balancing Yin and Yang, thus healing and improving the internal immune ability of the body (30). To some extent, TCM has advantages in treating BC that modern medicine does not.
RUT has a wide range of physiological and pharmacological roles, including cardiac protection, lowering blood pressure, vasodilation, gastric mucosa protection, and the treatment of ulcerative colitis (31). RUT also has anti-depressant, anti-obesity, anti-tumor, analgesic, and anti-inflammatory effects (32). Recently, a number of studies have shown that RUT plays obvious blocking roles in multiple cancer processes. For example, RUT has an anti-proliferative role in ovarian cancer cells (33), RUT also prevents the growth of prostate cancer cells by regulating the T helper type 1 (Th1)-polarized immune balance (7). In our study, we first found that RUT prevented proliferation and accelerated the cell-cycle arrest of BC cells. However, the regulatory mechanism of RUT in BC remained unclear.
MiRNAs, which are small single-stranded RNAs, are highly conserved (9). Extensive research has shown that the dysregulation of miRNA is relevant to multiple cancer processes (11). The expression of miRNA is stable and is an important part of the gene regulation network. The deviation of miRNA has been proven to be relevant in many diseases, including cardiovascular disease, diabetes, kidney disease, and cancer (34). Studies have shown that miR-149-3p has significant inhibitory roles in cancer cell metastasis in ovarian cancer, colorectal cancer, and non-small cell lung cancer (24). Additionally, research suggests that the downregulation of miR-149-3p is also correlated to tumor size, invasion, and lymph node metastasis in BC (15). In our study, we also discovered that RUT upregulates miR-149-3p, and the reduction of miR-149-3p reverses the inhibitory effect of RUT on proliferation, and the induction of cell-cycle arrest in BC cells. Thus, we showed that miR-149-3p is a regulatory gene of RUT in BC.
Tumor growth depends on the proliferation of tumor cells and the generation of tumor blood vessels. VEGF is the most crucial molecule for accelerating tumor angiogenesis and is also key in the growth and metastasis of cancer cells (35). VEGF mainly binds to VEGFR in a paracrine way in tumor cells to activate downstream pathways, thus inducing tumor angiogenesis (36). Studies have shown that the VEGF protein is highly expressed in malignant tumors, including BC, and is also related to tumor metastasis (37). Further, research has confirmed that RUT suppresses angiogenesis by the VEGFR2 (16). Our results also revealed that RUT suppressed the angiogenesis of BC cells by miR-149-3p.
Autophagy is a cellular metabolic process in which target substances in the cytoplasm are transported to lysosomes for degradation (38). Autophagy not only maintains cell homeostasis and physiological metabolism, but also facilitates apoptosis. Research has shown that autophagy plays a key role in multiple tumor processes, which might provide novel ideas for tumor research (39,40). Additionally, miRNA accelerates apoptosis by regulating autophagy, thus further improving the sensitivity of BC cells to radiotherapy and chemotherapy (41). Currently, the miRNAs known to regulate autophagy in BC include miR-142-3p (42), miR-27a (43), and miR-1910-3p (44). MiR-149-3p has also been confirmed to regulate autophagy in multiple studies (45). Similarly, we found that RUT induced the autophagy of BC cells by miR-149-3p.
Through bioinformatics prediction, we accidentally found that miR-149-3p potentially binds to the S100A4 promoter region, which suggests that S100A4 might be a targeted regulatory gene of miR-149-3p. S100A4 is a key member of the S100 protein family (46). It has been reported that S100A4 not only induces cell proliferation and metastasis, but also inhibits the apoptosis cascade, and the mechanism may be related to the binding of p53. P53 is a regulator of the cell cycle, DNA repair, and apoptosis. Additionally, S100A4 has also been confirmed to be involved in adhesion, angiogenesis, extracellular matrix remodeling, and the metastasis of cancer cells (47). Research has also shown that RUT prevents angiogenesis by the VEGFR2 (16). Thus, we hypothesized that S100A4 is related to BC progression, which may also be targeted by miR-149-3p. Notably, we discovered miR-149-3p directly targets S100A4, and S100A4 reverses the decrease of miR-149-3p on the metastasis of BC cells, which suggests that the miR-149-3p/S100A4 axis may participate in the RUT-mediated blocking of BC progression by regulating the proliferation, cell-cycle arrest, apoptosis, autophagy, and angiogenesis pathways.
Conclusions
This study first proved that RUT has a certain therapeutic effect on BC to prevent the malignant behaviors (such as proliferation and apoptosis, autophagy and angiogenesis), and its mechanism may be related to miR-149-3p/S100A4 axis. Therefore, we suggested that the RUT-mediated miR-149-3p/S100A4 axis might be the underlying therapeutic and prognostic targets for BC. However, there are some limitations to the current study. For example, more experiments are needed to verify the autophagy and angiogenesis of BC cells. Additionally, the in vivo research will also be supplemented in future research. Besides, the study of lncRNA regulating the miR-149-3p/S100A4 axis is also the focus of our future research. And the signaling pathway can also be further investigated and verified in the miR-149-3p/S100A4-mediated BC process. According to literature reports (26,31,48-50), the research of RUT on various diseases (heart disease, ulcerative colitis, obesity, tumors) is still based on cell and animal models, and its therapeutic dose and effect on patients in clinical practice also need to be further explored.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3765/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3765/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3765/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. All patients who participated in this study signed an informed consent form, and this study was approved by the Institutional Ethics Board of Wuhan Asia General Hospital (No. WAGHMEC-KY-2022009). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Adv Exp Med Biol 2019;1152:51-64. [Crossref] [PubMed]
- Winters S, Martin C, Murphy D, et al. Breast Cancer Epidemiology, Prevention, and Screening. Prog Mol Biol Transl Sci 2017;151:1-32. [Crossref] [PubMed]
- Coughlin SS. Epidemiology of Breast Cancer in Women. Adv Exp Med Biol 2019;1152:9-29. [Crossref] [PubMed]
- Barzaman K, Karami J, Zarei Z, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol 2020;84:106535. [Crossref] [PubMed]
- Nagini S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anticancer Agents Med Chem 2017;17:152-63. [Crossref] [PubMed]
- Tian KM, Li JJ, Xu SW. Rutaecarpine: A promising cardiovascular protective alkaloid from Evodia rutaecarpa (Wu Zhu Yu). Pharmacol Res 2019;141:541-50. [Crossref] [PubMed]
- Lin JY, Yeh TH. Rutaecarpine administration inhibits cancer cell growth in allogenic TRAMP-C1 prostate cancer mice correlating with immune balance in vivo. Biomed Pharmacother 2021;139:111648. Erratum in: Biomed Pharmacother 2021;141:111751. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. [Crossref] [PubMed]
- Xiong Q, Su H. MiR-325-3p functions as a suppressor miRNA and inhibits the proliferation and metastasis of glioma through targeting FOXM1. J Integr Neurosci 2021;20:1019-28. [Crossref] [PubMed]
- Cai WL, Huang WD, Li B, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer 2018;17:9. [Crossref] [PubMed]
- Qianqian Tang SW, Qiao X, Wang F, et al. MiR-29 promotes ovarian carcinoma cell proliferation through the PTEN pathway. Eur J Gynaecol Oncol 2020;41:774-8. [Crossref]
- Liu Q, Liu S, Wang D. Overexpression of microRNA-21 decreased the sensitivity of advanced cervical cancer to chemoradiotherapy through SMAD7. Anticancer Drugs 2020;31:272-81. [Crossref] [PubMed]
- Yang D, Du G, Xu A, et al. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res 2017;7:2209-19. [PubMed]
- Jiang Z, Ma Y, Tian T, et al. Maimendong and Qianjinweijing Tang (Jin formula) suppresses lung cancer by regulation of miR-149-3p. J Ethnopharmacol 2020;258:112836. [Crossref] [PubMed]
- Zhang M, Gao D, Shi Y, et al. miR-149-3p reverses CD8+ T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol 2019;9:190061. [Crossref] [PubMed]
- Ji L, Wu M, Li Z. Rutacecarpine Inhibits Angiogenesis by Targeting the VEGFR2 and VEGFR2-Mediated Akt/mTOR/p70s6k Signaling Pathway. Molecules 2018;23:2047. [Crossref] [PubMed]
- Gao YX, Jiang LL, Zhang Q, et al. Rutaecarpine protects against bleomycin-induced pulmonary fibrosis through inhibiting Notch1/eIF3a signaling pathway in rats. Zhongguo Zhong Yao Za Zhi 2018;43:3530-8. [PubMed]
- Zeng SY, Yang L, Lu HQ, et al. Rutaecarpine prevents hypertensive cardiac hypertrophy involving the inhibition of Nox4-ROS-ADAM17 pathway. J Cell Mol Med 2019;23:4196-207. [Crossref] [PubMed]
- Peng WJ, Liu Y, Yu YR, et al. Rutaecarpine prevented dysfunction of endothelial gap junction induced by Ox-LDL via activation of TRPV1. Eur J Pharmacol 2015;756:8-14. [Crossref] [PubMed]
- Ma J, Chen L, Fan J, et al. Dual-targeting Rutaecarpine-NO donor hybrids as novel anti-hypertensive agents by promoting release of CGRP. Eur J Med Chem 2019;168:146-53. [Crossref] [PubMed]
- Surbala L, Singh CB, Devi RV, et al. Rutaecarpine exhibits anti-diabetic potential in high fat diet-multiple low dose streptozotocin induced type 2 diabetic mice and in vitro by modulating hepatic glucose homeostasis. J Pharmacol Sci 2020;143:307-14. [Crossref] [PubMed]
- Kim SB, Chae GW, Lee J, et al. Activated Notch1 interacts with p53 to inhibit its phosphorylation and transactivation. Cell Death Differ 2007;14:982-91. [Crossref] [PubMed]
- Zhan R, Xu K, Pan J, et al. Long noncoding RNA MEG3 mediated angiogenesis after cerebral infarction through regulating p53/NOX4 axis. Biochem Biophys Res Commun 2017;490:700-6. [Crossref] [PubMed]
- Liang Y, Hou L, Li L, et al. Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene 2020;39:469-85. [Crossref] [PubMed]
- Zou T, Zeng C, Qu J, et al. Rutaecarpine Increases Anticancer Drug Sensitivity in Drug-Resistant Cells through MARCH8-Dependent ABCB1 Degradation. Biomedicines 2021;9:1143. [Crossref] [PubMed]
- Zhang Y, Yan T, Sun D, et al. Rutaecarpine inhibits KEAP1-NRF2 interaction to activate NRF2 and ameliorate dextran sulfate sodium-induced colitis. Free Radic Biol Med 2020;148:33-41. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Xu M, Li Y, Li W, et al. Immune and Stroma Related Genes in Breast Cancer: A Comprehensive Analysis of Tumor Microenvironment Based on the Cancer Genome Atlas (TCGA) Database. Front Med (Lausanne) 2020;7:64. [Crossref] [PubMed]
- Maughan KL, Lutterbie MA, Ham PS. Treatment of breast cancer. Am Fam Physician 2010;81:1339-46. [PubMed]
- Qi F, Zhao L, Zhou A, et al. The advantages of using traditional Chinese medicine as an adjunctive therapy in the whole course of cancer treatment instead of only terminal stage of cancer. Biosci Trends 2015;9:16-34. [Crossref] [PubMed]
- Zhao B, Wang Y, Liu R, et al. Rutaecarpine Ameliorated High Sucrose-Induced Alzheimer's Disease Like Pathological and Cognitive Impairments in Mice. Rejuvenation Res 2021;24:181-90. [Crossref] [PubMed]
- Choi JH, Jin SW, Lee GH, et al. Rutaecarpine Protects against Acetaminophen-Induced Acute Liver Injury in Mice by Activating Antioxidant Enzymes. Antioxidants (Basel) 2021;10:86. [Crossref] [PubMed]
- Yu CH, Lin RC, Wang PS. Anti-Proliferative Effects of Evodiamine and Rutaecarpine on Human Ovarian Cancer Cell Line SKOV3. Biol Reprod 2010;83:134. [Crossref]
- Tiwari A, Mukherjee B, Dixit M. MicroRNA Key to Angiogenesis Regulation: MiRNA Biology and Therapy. Curr Cancer Drug Targets 2018;18:266-77. [Crossref] [PubMed]
- Itatani Y, Kawada K, Yamamoto T, et al. Resistance to Anti-Angiogenic Therapy in Cancer-Alterations to Anti-VEGF Pathway. Int J Mol Sci 2018;19:1232. [Crossref] [PubMed]
- Frezzetti D, Gallo M, Maiello MR, et al. VEGF as a potential target in lung cancer. Expert Opin Ther Targets 2017;21:959-66. [Crossref] [PubMed]
- Lin X, Khalid S, Qureshi MZ, et al. VEGF mediated signaling in oral cancer. Cell Mol Biol (Noisy-le-grand) 2016;62:64-8. [Crossref] [PubMed]
- Ravanan P, Srikumar IF, Talwar P. Autophagy: The spotlight for cellular stress responses. Life Sci 2017;188:53-67. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer 2017;36:52. [Crossref] [PubMed]
- Wen N, Lv Q, Du ZG. MicroRNAs involved in drug resistance of breast cancer by regulating autophagy. J Zhejiang Univ Sci B 2020;21:690-702. [Crossref] [PubMed]
- Liang L, Fu J, Wang S, et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B 2020;10:1036-46. [Crossref] [PubMed]
- Ueda S, Takanashi M, Sudo K, et al. miR-27a ameliorates chemoresistance of breast cancer cells by disruption of reactive oxygen species homeostasis and impairment of autophagy. Lab Invest 2020;100:863-73. [Crossref] [PubMed]
- Wang B, Mao JH, Wang BY, et al. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett 2020;489:87-99. [Crossref] [PubMed]
- Wang N, Zhou P, Chen Y, et al. MicroRNA-149: A review of its role in digestive system cancers. Pathol Res Pract 2020;216:153266. [Crossref] [PubMed]
- Serrano A, Apolloni S, Rossi S, et al. The S100A4 Transcriptional Inhibitor Niclosamide Reduces Pro-Inflammatory and Migratory Phenotypes of Microglia: Implications for Amyotrophic Lateral Sclerosis. Cells 2019;8:1261. [Crossref] [PubMed]
- Fei F, Qu J, Zhang M, et al. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 2017;8:73219-39. [Crossref] [PubMed]
- Estari RK, Dong J, Chan WK, et al. Time effect of rutaecarpine on caffeine pharmacokinetics in rats. Biochem Biophys Rep 2021;28:101121. [Crossref] [PubMed]
- Song XM, Li BJ, Zhang YY, et al. Rutaecarpine enhances the anti-diabetic activity and hepatic distribution of metformin via up-regulation of Oct1 in diabetic rats. Xenobiotica 2021;51:818-30. [Crossref] [PubMed]
- Chen D, Duan Y, Yu S, et al. Rutaecarpine Promotes Adipose Thermogenesis and Protects against HFD-Induced Obesity via AMPK/PGC-1α Pathway. Pharmaceuticals (Basel) 2022;15:469. [Crossref] [PubMed]


