Biomarkers of risk to develop lung cancer in the new screening era
Introduction: why develop new biomarkers for early detection of lung cancer?
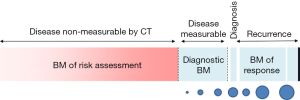
The rationale for developing biomarkers for the early detection of lung cancer is very strong and well established. It stems from the fact that, at the population level, the earlier we detect the disease, the better the outcome and the lower the health care cost. The impetus for biomarker development has grown stronger since the NLST trial demonstrated that early detection via chest CT screening reduced the relative risk for lung cancer death in the high risk individuals (1). Low dose chest CT in this group alone may save up to 12,000 lives a year, but it represents only about 8% of individuals dying of this disease every year. Thus, much is to be done to capture these lung cancers that escape chest CT screening as currently recommended despite its high sensitivity and specificity (2). The reason for limited detection relates to how many at-risk individuals are studied with CT and to how we best define this risk. Detection and careful management of indeterminate pulmonary nodules are integral parts of this effort. Lung cancer screening using chest CT also raises many questions, some of which could be addressed with well poised biomarkers. For example, who is at utmost risk for lung cancer? How do we expand the screening criteria from the NLST without causing more harm than good? Once the CT screening studies are done, how do we approach a non-invasive diagnosis of lung cancer? How do we prevent the overdiagnosis bias? In a previous report, we discussed the difference between biomarkers of risk for developing disease and diagnostic biomarkers (3). Here we focus on biomarkers that could be used in a risk assessment evaluation for screening programs (Figure 1). We do not discuss diagnostic biomarkers, predictive biomarkers of disease behavior, or biomarkers that could be used as intermediate endpoint for chemoprevention.
An ideal biomarker to assess the risk of developing lung cancer
Biomarkers are usually understood as molecular entities quantifiable in biological specimens. An ideal biomarker would be easily measurable (of sufficient abundance), accurate, quantitative, reproducible, biological plausible, cheap and adopted by practitioners to modify the management of high risk individuals (4). None of the candidate molecular biomarkers have demonstrated thus far reduction in lung cancer mortality. Regardless, there is tremendous growth in biomarker research because of the incredible potential it would offer. In this context, we are looking for biomarkers that are typically better at providing strong positive predictive value (PPV) (usually with great specificity) or negative predictive value (NPV) (usually with greater sensitivity). The ideal biomarker will provide insights into the management of high risk individuals. Several investigators have developed models to predict individual’s risk of developing lung cancer (5-15). These models differ in the number and type of predictor variables and most models do not include molecular biomarkers. To date, no single model is systematically applied to high-risk individuals as a screening tool. Yet CMS recommends the use of decision aids as part of shared decision-making during the required visit prior to screening (16). None of the decision aids have included molecular biomarkers, and of those have made the guidelines recommendations for Components Necessary for High-Quality Lung Cancer Screening (17) neither in the ATS ACCP guidelines for implementation (16). Biomarkers are likely to get there as evidence for clinical utility is being tested. The evidence will include stage shift, added value to existing clinical tools, cost effectiveness and hopefully cancer control.
Biomarkers of risk of developing lung cancer
In this section we will discuss current molecular biomarkers of risk assessment in those without measurable disease and before a chest CT has been done. Consideration of the use of such biomarkers should trigger a discussion with the patient before ordering it to address the intent of the test and the implications of the possible results. Many biomarkers have been developed over the years to predict tumor development (18).
Let us consider the characteristics of such a biomarker to assess the risk of lung cancer. For screening purposes, given the low prevalence of disease, a strong NPV of a test is a very attractive feature. Thus, high sensitivity with low false negative could rule out disease and reassure individuals. In this respect a negative chest CT provides great risk reduction (19).
Some biomarkers may be quite sensitive at the risk of overcalling the disease. This is common with inflammatory markers and miRNAs, which are typically elevated during pathogenesis and yet not associated with measurable disease. These markers are associated with increased odds of developing the disease at five years but with relatively small odds ratios. But it is unknown at what odds ratios it is worth incorporating these biomarkers into screening programs remains unknown.
Some serum-based inflammation marker levels are associated with prospective lung cancer risk. A panel of 11 proteins was associated with lung cancer risk in the PLCO trial where four markers (CRP, BCA-1/CXCL13, MDC/CCL22, and IL-1RA) provided an overall estimated 10-year cumulative risks of lung cancer of 0.16% in never smokers, 1.8% in former smokers, and 5.0% in current smokers. Such a profile is a good candidate for validation in other cohorts and registry studies (20).
Circulating miRNA profiles are associated with malignancy, including lung cancer (21). They also are stable in blood despite high levels of circulating RNases, making them prime candidates to serve as biomarkers of disease (22,23). Although the strength of these signatures seems to reside in the diagnostic setting (24-26) by potentially reducing the need for additional diagnostic evaluation, it remains to be determined whether these may add value to a true risk assessment strategy.
High specificity on the other hand is always desirable so we do not overcall cancers (false positive). Should such a test be positive, it would push individuals into a higher risk group to consider appropriate surveillance. A few examples of such candidates include autoantibodies, epithelial chromosomal imbalances, and cfDNA, and they are summarized here.
Autoantibodies to tumor associated antigens have been found to precede clinical presentation s of cancer by months to years (27-29), and they may actually circulate before the disease is measurable on CT of the chest (30-33). The difficulty with using autoantibodies as a screening tool is that the sensitivity is around 40%, thereby missing quite a few cancers. A positive test may be useful as an adjunct to detection of the disease by CT or bronchoscopy and could inform the decision of surveillance versus further intervention. By using a panel of antigens, autoantibodies can be detectable 1–5 years before detection of lung cancer on incidence screening (27). The robust specificity of this approach indicates that autoantibody panels may make a significant contribution in the future to the diagnosis and screening of individuals at risk for lung cancer. As a screen, such autoantibody test may be more appropriate for populations at high risk for lung cancer.
Chromosomal imbalances have been tested in the sputum of high risk individuals and provide a candidate biomarker of tumor development (34,35). Using probes to EGFR, MYC, 5p15, and CEP 6 by multicolor FISH, Varella-Garcia et al. demonstrated CA-FISH as a potential marker for incident lung cancer (36). Whether this approach may assess the lung cancer risk of high risk individuals in a screening context needs to be demonstrated.
Another example of a high specificity test is circulating tumor DNA (ctDNA). The presence of cell free DNA circulating in plasma or serum has been described in patients with cancer (37,38). The analysis of ctDNA may give valuable information about the underlying genomic alterations of individual’s tumor. Although the precise mechanism of DNA release into the blood remains unknown, it is likely to be derived from apoptotic and necrotic tumor cells. ctDNA is exquisitely specific but its sensitivity is limited by the number of circulating molecules, and as a screening tool suffers the risk of missing too many cancers (39). Such markers, however, could be helpful when positive and provide diagnostic and prognostic information. The diagnostic accuracy of quantitative analysis of circulating tumor DNA is not very different than conventional serum biomarkers for lung cancer screening. Yet the sensitivity of the assay remains the major limitation of the test even in stage 1 lung cancer. A multigene panel analysis of ctDNA may lead to increased sensitivity, but for now this marker is more likely to apply to a diagnostic setting.
What would such biomarker of risk really measure?
The biomarker could measure a genetic risk (e.g., altered metabolism of carcinogens, DNA repair machinery abnormalities, predisposition to inflammation, or germline mutations) or the influence of the environment on tumor development (exposure to carcinogens or surrogates of risk such as epigenetic changes in the airway epithelium or the prevalence of preinvasive lesions). There has been recent interest in the potential for genetic variants to give insight into the pathogenesis of lung cancer. These variants indicate that there is great heterogeneity in mechanisms of disease development that is modulated by inherited genetic variation (40). With these come the opportunity to improve models predicting lung cancer risk. SNP genotype signature data may add value to the performance of clinical variables for risk prediction by re-assigning risk in 26% of the screening participants (41). Yet most risk assessment predictive models have shown little improvement with the addition of genetic factors (42-44). While genetics are not yet incorporated into lung cancer malignancy prediction models beside a family history of lung cancer, it is likely that a profile of many genetic markers will be necessary to be clinically useful biomarkers for risk assessment. Future development of predictive models will incorporate previously identified predictors and newly identified biomarkers, genetic or otherwise.
What is the metric for success of these biomarkers of risk assessment?
A critical goal of biomarker research is to add value to existing risk assessment standards, and the biomarker should be designed to supplement the current diagnostic/management tools (45). The biomarker of interest for risk assessment for lung cancer should therefore provide added value to the clinical or risk models such as the PanCan, Bach or Spitz models. Odds ratios and post-test probabilities are metrics of most relevance for clinical practice, but these metrics are often not sufficient (46,47). A good predictive value of any biomarker or test result, by itself, is no guarantee for relevant added predictive value when combined with the standard predictors. Comparing area under the ROC curves, testing for the net reclassification improvement [how many times a sample is now reclassified in the correct group of cancer vs. no cancer (48) for categorical variables], or using the integrated discrimination improvement (IDI) for continuous variables may provide other valuable metrics of success. The positive likelihood ratio (PLR) indicates how much the odds of the disease increase when a test is positive, and the negative likelihood ratio (NLR) indicates how much the odds of the disease decrease when a test is negative. Likelihood ratios of >10 or <0.1 generate often conclusive changes from pretest to posttest probability. The ultimate metric of success for biomarker of risk in a screening program, however, is to reduce the number needed to screen and to increase the number of cancers found at an early stage.
Reasons for failure
Some of the reasons why the field has failed to deliver greater number of effective biomarkers of risk are discussed above in the section describing an ideal biomarker. The lack of analytical reproducibility or of biological variability is a common obstacle. Attempts to fit the biomarker with characteristics that are not inherently strong also often lead to lack of validation. Appropriate study design is crucial for the derivation of a potential biomarker for screening much like the successful validation of a promising biomarker for clinical use. Derivation of a biomarker useful for lung cancer screening may be more successful using a nested case-control study design within a prospective longitudinal cohort following the PRoBE design (49). These are difficult studies to design in some biological specimens given the limited number of prospective cohorts meeting the tissue collection required for the analytical method of interest, such as the analysis of RNA in airway specimens (bronchial or nasal) or volatile organic compounds in the exhaled breath. Many candidate biomarkers have been developed in a case control study design. Most of them have failed when applied to risk assessment because of the lack of sensitivity. One common mistake is to ignore the inherent characteristics of the biomarker and apply it to a clinical situation outside of the context in which it was designed.
Areas to explore
There is a clear need to evaluate the benefit of risk assessment biomarkers with repeated measures over time. The assumption is that as risk increases, molecular moieties should be more readily available (e.g., in the circulation) over time. This may be true for tumor specific antigens and ctDNA, but would not apply to genetic risk. Statistical models could test the ability of different biomarkers to complement each other in a single population, in order to eventually determine those that could be tested prospectively. Given biomarkers’ non-specificity and commonality in predicting diseases, modeling multiple markers of the same clinical diagnostic criteria can be used to develop more accurate individual and cumulative risk estimates for specific diseases. We should therefore consider a joint effects approach to determine individual biomarker associations as well as to ascertain the impact of simultaneous increases in multiple biomarker concentrations on the diagnosis of lung cancer. Biomarkers of risk would ideally be tested prospectively in a randomized clinical trial. However, given the relatively low prevalence of this disease, the number needed to screen may be prohibitive; therefore the development of registries is most appropriate. Registries are longitudinal cohort prospective studies where a biomarker is introduced but does not force providers to change their management. The lead time to diagnosis may be sufficient to cause a stage shift and therefore improve outcome. The discovery of other traits associated with lung cancer using PheWAS studies may be of relevance to the field in terms of increasing our ability to refine the at-risk population (50-53). Finally, it is through better understanding of the biology of cancer development and of preinvasive lesions that we will shed further light into the field of biomarker research.
Acknowledgements
Funding: The work was supported by the National Institutes of Health (CA152662 and CA186145 to PPM).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- van Klaveren RJ, Oudkerk M, Prokop M, et al. Management of lung nodules detected by volume CT scanning. N Engl J Med 2009;361:2221-9. [Crossref] [PubMed]
- Massion PP, Walker RC. Indeterminate pulmonary nodules: risk for having or for developing lung cancer? Cancer Prev Res (Phila) 2014;7:1173-8. [Crossref] [PubMed]
- Srivastava S, Gopal-Srivastava R. Biomarkers in cancer screening: a public health perspective. J Nutr 2002;132:2471S-2475S. [PubMed]
- Bach PB, Kattan MW, Thornquist MD, et al. Variations in lung cancer risk among smokers. J Natl Cancer Inst 2003;95:470-8. [Crossref] [PubMed]
- Cassidy A, Myles JP, van Tongeren M, et al. The LLP risk model: an individual risk prediction model for lung cancer. Br J Cancer 2008;98:270-6. [Crossref] [PubMed]
- Peto R, Darby S, Deo H, et al. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case-control studies. BMJ 2000;321:323-9. [Crossref] [PubMed]
- Prindiville SA, Byers T, Hirsch FR, et al. Sputum cytological atypia as a predictor of incident lung cancer in a cohort of heavy smokers with airflow obstruction. Cancer Epidemiol Biomarkers Prev 2003;12:987-93. [PubMed]
- Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst 2007;99:715-26. [Crossref] [PubMed]
- Tammemagi MC, Freedman MT, Pinsky PF, et al. Prediction of true positive lung cancers in individuals with abnormal suspicious chest radiographs: a prostate, lung, colorectal, and ovarian cancer screening trial study. J Thorac Oncol 2009;4:710-21. [Crossref] [PubMed]
- Tammemagi CM, Pinsky PF, Caporaso NE, et al. Lung cancer risk prediction: Prostate, Lung, Colorectal And Ovarian Cancer Screening Trial models and validation. J Natl Cancer Inst 2011;103:1058-68. [Crossref] [PubMed]
- Kovalchik SA, Tammemagi M, Berg CD, et al. Targeting of low-dose CT screening according to the risk of lung-cancer death. N Engl J Med 2013;369:245-54. [Crossref] [PubMed]
- Cronin KA, Gail MH, Zou Z, et al. Validation of a model of lung cancer risk prediction among smokers. J Natl Cancer Inst 2006;98:637-40. [Crossref] [PubMed]
- McRonald FE, Yadegarfar G, Baldwin DR, et al. The UK Lung Screen (UKLS): demographic profile of first 88,897 approaches provides recommendations for population screening. Cancer Prev Res (Phila) 2014;7:362-71. [Crossref] [PubMed]
- Marcus MW, Chen Y, Raji OY, et al. LLPi: Liverpool Lung Project Risk Prediction Model for Lung Cancer Incidence. Cancer Prev Res (Phila) 2015;8:570-5. [Crossref] [PubMed]
- Wiener RS, Gould MK, Arenberg DA, et al. An official American Thoracic Society/American College of Chest Physicians policy statement: implementation of low-dose computed tomography lung cancer screening programs in clinical practice. Am J Respir Crit Care Med 2015;192:881-91. [Crossref] [PubMed]
- Mazzone P, Powell CA, Arenberg D, et al. Components necessary for high-quality lung cancer screening: American College of Chest Physicians and American Thoracic Society Policy Statement. Chest 2015;147:295-303. [Crossref] [PubMed]
- Hassanein M, Callison JC, Callaway-Lane C, et al. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992-1006. [Crossref] [PubMed]
- Maisonneuve P, Bagnardi V, Bellomi M, et al. Lung cancer risk prediction to select smokers for screening CT--a model based on the Italian COSMOS trial. Cancer Prev Res (Phila) 2011;4:1778-89. [Crossref] [PubMed]
- Pine SR, Mechanic LE, Enewold L, et al. Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst 2011;103:1112-22. [Crossref] [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [Crossref] [PubMed]
- Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 2011;108:3713-8. [Crossref] [PubMed]
- Bianchi F, Nicassio F, Marzi M, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503. [Crossref] [PubMed]
- Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768-73. [Crossref] [PubMed]
- Montani F, Marzi MJ, Dezi F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063. [Crossref] [PubMed]
- Wozniak MB, Scelo G, Muller DC, et al. Circulating MicroRNAs as Non-Invasive Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. PLoS One 2015;10:e0125026. [Crossref] [PubMed]
- Zhong L, Coe SP, Stromberg AJ, et al. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol 2006;1:513-9. [Crossref] [PubMed]
- Trivers GE, De Benedetti VM, Cawley HL, et al. Anti-p53 antibodies in sera from patients with chronic obstructive pulmonary disease can predate a diagnosis of cancer. Clin Cancer Res 1996;2:1767-75. [PubMed]
- Desmetz C, Bascoul-Mollevi C, Rochaix P, et al. Identification of a new panel of serum autoantibodies associated with the presence of in situ carcinoma of the breast in younger women. Clin Cancer Res 2009;15:4733-41. [Crossref] [PubMed]
- Chapman CJ, Murray A, McElveen JE, et al. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 2008;63:228-33. [Crossref] [PubMed]
- Lam S, Boyle P, Healey GF, et al. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011;4:1126-34. [Crossref] [PubMed]
- Chapman CJ, Thorpe AJ, Murray A, et al. Immunobiomarkers in small cell lung cancer: potential early cancer signals. Clin Cancer Res 2011;17:1474-80. [Crossref] [PubMed]
- Pedchenko T, Mernaugh R, Parekh D, et al. Early detection of NSCLC with scFv selected against IgM autoantibody. PLoS One 2013;8:e60934. [Crossref] [PubMed]
- Varella-Garcia M, Chen L, Powell RL, et al. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med 2007;176:505-12. [Crossref] [PubMed]
- Varella-Garcia M, Kittelson J, Schulte AP, et al. Multi-target interphase fluorescence in situ hybridization assay increases sensitivity of sputum cytology as a predictor of lung cancer. Cancer Detect Prev 2004;28:244-51. [Crossref] [PubMed]
- Varella-Garcia M, Schulte AP, Wolf HJ, et al. The detection of chromosomal aneusomy by fluorescence in situ hybridization in sputum predicts lung cancer incidence. Cancer Prev Res (Phila) 2010;3:447-53. [Crossref] [PubMed]
- Teare MD, Woll PJ. Genomic tests: unreliable for cancer? A focus on circulating DNA and lung cancer. Expert Rev Mol Diagn 2007;7:699-702. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet 2012;21:4980-95. [Crossref] [PubMed]
- Young RP, Hopkins RJ, Gamble GD. Clinical applications of gene-based risk prediction for lung cancer and the central role of chronic obstructive pulmonary disease. Front Genet 2012;3:210. [Crossref] [PubMed]
- Spitz MR, Etzel CJ, Dong Q, et al. An expanded risk prediction model for lung cancer. Cancer Prev Res (Phila) 2008;1:250-4. [Crossref] [PubMed]
- Raji OY, Agbaje OF, Duffy SW, et al. Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: the Liverpool Lung Project. Cancer Prev Res (Phila) 2010;3:664-9. [Crossref] [PubMed]
- Young RP, Hopkins RJ, Hay BA, et al. Lung cancer susceptibility model based on age, family history and genetic variants. PLoS One 2009;4:e5302. [Crossref] [PubMed]
- Hendriksen JM, Geersing GJ, Moons KG, et al. Diagnostic and prognostic prediction models. J Thromb Haemost 2013;11 Suppl 1:129-41. [Crossref] [PubMed]
- Moons KG. Criteria for scientific evaluation of novel markers: a perspective. Clin Chem 2010;56:537-41. [Crossref] [PubMed]
- Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 2007;115:928-35. [Crossref] [PubMed]
- Pencina MJ, D'Agostino RB Sr, D'Agostino RB Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157-72; discussion 207-12. [Crossref] [PubMed]
- Pepe MS, Feng Z, Janes H, et al. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 2008;100:1432-8. [Crossref] [PubMed]
- Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 2016;17:129-45. [Crossref] [PubMed]
- Wang X, Pandey AK, Mulligan MK, et al. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun 2016;7:10464. [Crossref] [PubMed]
- Gamazon ER, Wheeler HE, Shah KP, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 2015;47:1091-8. [Crossref] [PubMed]
- Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med 2015;7:41. [Crossref] [PubMed]


