
SMC4 enhances the chemoresistance of hepatoma cells by promoting autophagy
Introduction
In 2020, primary liver cancer was the sixth most commonly diagnosed cancer and the third leading cause of cancer-related death worldwide, with approximately 906,000 new cases and 830,000 liver cancer-related deaths (1). The most common type of primary liver cancer is hepatocellular carcinoma (HCC), which accounts for 80–90% of cases (2). Globally, the incidence of HCC is highest within Asia and Africa, with the two regions comprising around 80% of all HCC cases worldwide (3). Significant disparities in the incidence and outcomes of HCC exist even within more developed countries due to socioeconomic factors such as race/ethnicity (4). Chronic viral hepatitis is one of the most important risk factors for HCC; in 2019, 69% of HCC cases were identified in association with hepatitis B virus (HBV) or hepatitis C virus (HCV) (5,6).
However, the incidence and cancer-specific mortality rates continue to increase in many countries, and the majority of HCC patients still present at an advanced stage in many parts of the world (7,8). At present, the treatment of HCC is a comprehensive approach based on surgical resection, which has resulted in a 5-year overall survival (OS) rate of 60–70% (9,10). Unfortunately, drug therapy agents are urgently needed, as more than half of HCC patients are already at an intermediate or advanced stage of disease when they are first diagnosed. Owing to the genetic, metabolic, and inflammatory heterogeneity of liver cancer, the development of therapies is challenging (11). Common therapeutic drugs include 5-fluorouracil (5-FU), cisplatin, gemcitabine, doxorubicin, sorafenib, regorafenib, lenvatinib, programmed cell death protein 1 (PD-1), etc., which have partially prolonged the survival rate of advanced HCC patients (12-15). Given that their efficacy is quite limited, research on the resistance of hepatoma cells to 5-FU and other chemotherapeutic drugs is urgently needed. A study has shown that autophagy plays an important role in the resistance of HCC cells to chemotherapy drugs (16). Therefore, it is of great clinical value to explore the mechanism of autophagy in improving the sensitivity of hepatocellular carcinoma to chemotherapy drugs.
In our previous study, we demonstrated that structural maintenance of chromosome 4 (SMC4) is related to the progression and invasion of hepatoma cells (17). We identified a novel miR-219/SMC4/JAK2/Stat3 signaling pathway whose function may contribute to the development of HCC (18). In the present study, we aim to provide new insight into the function of SMC4 in the development of chemotherapeutic drug resistance in hepatoma cells and the underlying mechanisms. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3623/rc).
Methods
Cell culture and establishment of drug-resistant cell lines
The human hepatoma cell lines 97-H, HepG2, Bel-7405, and SMMC-7721 were used in this study and were purchased from the Chinese Academy of Sciences (Shanghai, China). The cells were grown in complete Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 (carbon dioxide) at 37 °C. Different concentrations of 5-FU (0, 50, 100, 200, 400, 800, and 1,600 µM) were added, and the cells were incubated for 48 h. The cell survival rate was tested using the Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China) assay, and the half maximal inhibitory concentration (IC50) value was calculated by the SPSS 20.0. The IC50 value was used to generate 5-FU-resistant hepatoma cell lines in the culture.
CCK-8
The proliferative ability of the cells was measured using the CCK-8 (Beyotime). The cells were seeded in 96-well plates at a density of 10,000 cells per well in 100 µL per well. The cells were allowed to adhere for 48 h, after which, CCK-8 solution was added and incubated at different time points, and the cells were incubated in an incubator for 1 h. Cell proliferation was measured based on the absorbance at 450 nm using a BioTek Synergy HT plate reader (Winooski, VT, USA).
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen, Carlsbad, California, USA) and reverse-transcribed by an RT-PCR kit (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was then carried out with the SYBR Green PCR Master Mix (Invitrogen) in a real-time PCR system (Applied Biosystems, Carlsbad, California, USA) according to a standard quantitative PCR procedure. All reactions were performed in triplicate, and reactions without reverse transcriptase were used as negative controls. All of the primer sequences for SMC4 are summarized in Table 1. β-actin and U6 (Sangon Biotech, Shanghai, China) were used as the controls.
Table 1
| Primer | Sequence |
|---|---|
| SMC4-F | 5'-GAGAAAATTCTGGGACCTTT-3' |
| SMC4-R | 5'-TCTGAATGTCCTTGTGTTCA-3' |
| Homo-GAPDH-F | 5'-TGCACCACCAACTGCTTAGC-3' |
| Homo-GAPDH-R | 5'-GGCATGGACTGTGGTCATGAG-3' |
| lncRNA-XIST-F | 5'-GCATAACTCGGCTTAGGGCT-3' |
| lncRNA-XIST-R | 5'-TCCTCTGCCTGACCTGCTAT-3' |
| β-actin-F | 5'-ACCCTGAAGTACCCCATCGAG-3' |
| β-actin-R | 5'-AGCACAGCCTGGATAGCAAC-3' |
| miR-miR-219a-F | 5'-ACACTCCAGCTGGGTGATTGTCCAAACGC-3' |
| miR-miR-219a-R | 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGAATTGC-3' |
| U6snRNA-F | 5'-CTCGCTTCGGCAGCACA-3' |
| U6snRNA-R | 5'-AACGCTTCACGAATTTGCGT-3' |
Western blotting
Total protein was extracted from the cultured cells using a total protein extraction kit (ProMab Biotechnologies, Richmond, USA) according to the manufacturer’s instructions. The proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes. The membranes were incubated with the appropriate antibodies and dilutions and exposed to X-ray films to visualize the positive protein bands. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnologies, CA, USA)/β-actin (Santa Cruz Biotechnologies, CA, USA) was used as the control.
Vector construction and transfection
For the knockdown experiments, the cells were seeded in six-well plates and transfected with siRNA (small interfering RNA) using the Lipofectamine RNAiMAX transfection reagent (GenePharma, Shanghai, China) according to the manufacturer’s instructions. A study has shown that SMC4-Homo-830 greatly decreases the expression of SMC4 (18). SMC4-Homo-830 was cloned into the pGPU/GFP/Neo-shNC (RNAi control vector containing GFP gene, and includes neomycin resistant gene) vector (GenePharma) and transfected into the cell lines.
Immunofluorescence
The cells were washed with phosphate buffered saline (PBS) for 5 min and fixed for 20 min at 37 °C. Next, the cells were blocked with 5% goat serum for 20 min and incubated with a light chain 3 beta (LC3B) antibody (Abcam, Cambridge, UK) overnight at 4 °C. After washing three times in PBS, the cells were incubated with Alexa 488-conjugated goat anti-rabbit immunoglobulin G (IgG) (Santa Cruz Biotechnologies, Dallas, USA) for 40 min at 37 °C. After washing three times, the slides were immediately mounted in a mounting medium with DAPI [2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride]. The cells were then examined using a confocal laser-scanning microscope (LEICA TCS SP5, Germany), and the images were captured using Leica Confocal Software.
Flow cytometry assay
The indicated cells were harvested, washed with PBS-EDTA (ethylene diamine tetraacetic acid), and processed for apoptosis analysis using flow cytometry. Briefly, the fixed cells were centrifuged at 300 rpm for 10 min, washed with cold PBS, and then centrifuged again at 300 rpm for 10 min. Annexin V (100 µL) (GenePharma) was added to the cells and incubated for 15 min at room temperature. Cold 1× binding buffer (400 µL) was added, and the cells were analyzed by using a FACSCalibur instrument (BD Biosciences, Bedford, MA, USA) equipped with Cell Quest 3.3 software (BD Biosciences).
MTT (3-(4,5)-dimethylthiahiazo (-z-y1)-2,5-di-phenytetrazoliumromide) assay
The cells were added to 96-well plates at approximately 1×104 cells in 100 µL/well and cultured in a 37 °C and 5% CO2 cell incubator for 24 h. The appropriate concentration of drug was then added. Subsequently, the 96-well plates were incubated at 37 °C in air containing 5% CO2 and 100% humidity for the appropriate time. Next, 5× MTT (Abcam) was diluted to 1× MTT with diffusion buffer. Fifty microliters of 1× MTT were added to each well and incubated at 37 °C for 4 h. Thereafter, the supernatant was removed, 150 µL DMSO (dimethyl sulfoxide) was added to each well, and the plate was shaken. The optical density of each well was detected at 570 nm.
Luciferase reporter assay
The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to verify the interaction between the long non-coding RNA-XIST (lncRNA-XIST) promoter and miR-219. The miR-219 mimics and reporter gene plasmids were transfected with Lipofectamine™ 2000 (Thermo, Shanghai, China). The miR-219/Lipofectamine™ 2000 transfection complex was added to a 48-well plate containing cells and culture medium and shaken gently. The cell samples were then collected for lysis, and the luciferase activity was detected. Each group was analyzed in triplicate. When the Renilla luciferase was used as an internal reference, the firefly relative light unit (RLU) value was divided by the Renilla RLU value. The relative activation of target reporter genes among the different samples was compared based on the obtained ratio.
RNA pulldown assay
Bio-miR-219a-5p, Bio-miR-219a-5p (Mut, mutation), and Bio-miR-inhibitor NC were transfected into HepG2/HepG2/5-FU cells (80–90% confluence) for 48 h. Next, the cells were lysed, and the lysate was collected and incubated with magnetic beads (Sigma-Aldrich Chemical Company, MO, USA) combined with streptavidin. The protein-RNA complex absorbed by the beads was eluted. Total RNA was extracted using TRIzol, and the miR-219 expression was determined.
Invasion assay
The upper chamber of the transmembrane was coated with 50 mg/L Matrigel diluted (1:8) and dried at 4 °C. Diluted Matrigel (60–80 µL) was added to the polycarbonate film on the upper chamber (BD Biosciences) and allowed to polymerize to gel at 37 °C for 30 min. Next, the cells were digested and centrifuged, and the culture medium was discarded. Then, the cells were washed with PBS 1–2 times and resuspended in a serum-free medium containing bovine serum albumin (BSA). The cell density was adjusted to 5×104/mL. FBS-supplemented medium (1 mL) was added to the lower chamber of the plate (six wells) (Corning Incorporated, Corning, NY, USA). The cell suspension was added to the upper chamber. After culturing for an indicated duration, the cell membrane was extracted.
Wound healing assay
The wound healing assay was performed to measure the motility of cells. Firstly, the cell layer was wounded using a sterile micropipette tip. A total of 1.2×106 cells were seeded into 24-well plates and incubated for 24 h. After the cells became confluent, sterile 10-µL plastic pipette tips were used to draw a line-shaped scratch in the middle of the cell monolayer, and images were captured after the scratch was completed and used as a 0 h control. All of the cells were then washed with PBS three times. After being cultured at 37 °C in air containing 5% CO2, the cells in 24-well plate were extracted, and images were captured at the indicated time points.
Colony formation assay
Monolayers of cultured cells in the logarithmic growth phase were digested with 0.25% trypsin and then resuspended into single-cell suspensions and counted. The cells were seeded at a density of 500 cells per dish. The cells were incubated at 37 °C in air containing 5% CO2 for 2–3 weeks. The number of cells in each cell clone was counted, the cell colony formation rate was calculated, and images were captured.
Statistical analysis
Statistical analysis was carried out using SPSS 20.0 (Inc., Chicago, IL, USA) for Windows. The Student’s t-test was used to analyze the results expressed as the mean ± standard deviation (SD). Differences were considered significant when the P values were less than 0.05.
Results
Screening of hepatoma cell lines with high SMC4 expression
In our previous study (18), western blotting was performed to determine the expression of SMC4 in four hepatoma cell lines (SMMC-7721, Bel-7405, HepG2, and 97-H cells). The results showed that SMC4 expression was strongly upregulated in HepG2 cells compared to the other cells. Therefore, we chose HepG2 cells to generate a 5-FU-resistant hepatoma cell line and named it HepG2/5-FU.
Construction of a drug-resistant cell line
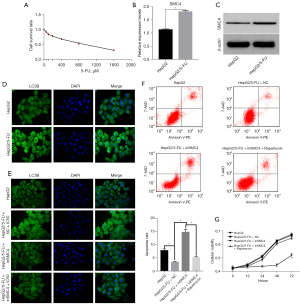
HepG2 cells were incubated with different concentrations of 5-FU (0, 50, 100, 200, 400, 800, and 1,600 µM). The CCK-8 assay was used to test the cell survival rate (Figure 1A). The results showed that with increasing 5-FU concentration, the proliferation rate of HepG2 cells decreased, suggesting that 5-FU can inhibit the proliferation activity of HepG2 cells. The IC50 value (785.9587) was calculated when the inhibition of cell proliferation reached 50%. The cells were then treated with the concentration until the drug-resistant cells were identified. The time required was approximately 3–4 weeks. At this time, the cells in the culture no longer died but rather proliferated in the presence of the drug, and thus, could be considered to have developed drug resistance.
The SMC4 expression level increased significantly in drug-resistant cell lines
RT-qPCR and western blotting were performed to confirm the expression level of SMC4 in drug-resistant cells (HepG2/5-FU) (Figure 1B,1C). The expression level of SMC4 in the HepG2/5-FU cell line was significantly increased compared to that in the HepG2 cell line. These results suggested that SMC4 might play an important role in the development of drug resistance in hepatoma cells.
SMC4 plays an important role in the autophagy in hepatoma cells
Previous studies have reported that tumor cells clear aging and degenerating cells by autophagy, preserve proliferating cells to adapt to the tumor environment, and maintain a high level of autophagy to induce drug resistance, which is an important mechanism of drug resistance in tumor cells (19,20). To further verify the role of SMC4 in autophagy in hepatoma cells, we first applied immunofluorescence to detect the level of LC3B in HepG2/5-FU cells to assess autophagy (21), and the results showed that the LC3B level in HepG2/5-FU cells was significantly increased compared to that in HepG2 cells (Figure 1D).
Second, an interference plasmid of SMC4 was cloned and stably transfected into the cell lines. Immunofluorescence assays were performed to evaluate the expression of LC3B in hepatoma cells (Figure 1E). The results showed that SMC4 intervention resulted in a decrease in autophagy in hepatoma cells compared to other treatments. The results also demonstrated that following SMC4 intervention, the level of autophagy was significantly reduced through the use of drugs, suggesting that the drug-resistant cells were resensitized to the drugs. Therefore, we concluded that SMC4 plays an important role in autophagy in hepatoma cells.
Finally, we used rapamycin to stimulate autophagy after SMC4 intervention. CCK-8 and flow cytometry assays were performed (Figure 1F,1G). In the experiment, group A was the control group and group B was the HepG2/5-FU + NC group. The expression of SMC4 in group B was higher than that in group A, so it was considered to have a higher autophagy level and stronger proliferative ability. Compared with group B, the knockdown of SMC4 in group C resulted in a decrease in the autophagy levels, suggesting that the level of autophagy was decreased and the level of apoptosis was increased in drug-resistant cell lines following SMC4 knockout. In group D, the autophagy activator rapamycin was used to induce drug-resistant cells to produce autophagy. Although SMC4 was knocked down, rapamycin still activated autophagy, which resulted in an increase in autophagy levels as well as a decrease in apoptosis and 5-FU-induced killing. It was further shown that autophagy was the key anti-drug factor and reduced the level of apoptosis.
Through competitive binding of miR-219, lncRNA-XIST effectively reduced the expression of SMC4, which led to the improvement of autophagy levels and drug resistance in hepatoma cells
A gene chip assay was performed to further study the upstream regulation of miR-219/SMC4. The results showed that the expression of two typical lncRNAs, lncRNA-XIST and lncRNA-TUG1, was upregulated (Figure 2A, Table 2). Secondly, database analysis (http://starbase.sysu.edu.cn/index.php) indicated that both lncRNA-XIST and lncRNA-TUG1 contain a binding site for miR-219, and further analysis showed that lncRNA-XIST and miR-219 have four binding sites (Table 3). Only the fourth site of lncRNA-XIST binding to miR-219 can competitively bind SMC4; therefore, we speculated that lncRNA-XIST might act as an intracellular miRNA sponge or regulatory molecule of miR-219.
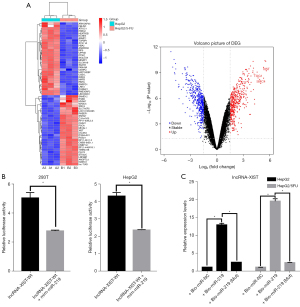
Table 2
| Name | Mir accession | Gene name | Target sites | Bio complex | Clip read Num | Cancer Num |
|---|---|---|---|---|---|---|
| hsa-miR-219a-5p | MIMAT0000276 | XIST | 4 | 8 | 66 | 1 |
| hsa-miR-219a-5p | MIMAT0000276 | SENP3-EIF4A1 | 1 | 5 | 126 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | MEG3 | 1 | 1 | 17 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | TUG1 | 1 | 4 | 976 | 11 |
| hsa-miR-219a-5p | MIMAT0000276 | CTD-2540L5.5 | 1 | 3 | 23 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | LINC00339 | 1 | 3 | 38 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | AP000783.2 | 1 | 1 | 8 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | RP6-24A23.7 | 1 | 4 | 0 | No data |
| hsa-miR-219a-5p | MIMAT0000276 | CTA-204B4.6 | 1 | 1 | 1 | No data |
Table 3
| Locus | Binding site |
|---|---|
| Locus 1 | miRNA-219: 3'-TCTTAACGCAAACCTGTTAGT-5' |
| Lnc-XIST: 5'-ACCCTTACATTTGGACAATCA-3' | |
| Locus 2 | miRNA-219: 3'-TCTTAACGCAAACCTGTTAGT-5' |
| Lnc-XIST: 5'-ACCCTTACATTTGGACAATCA-3' | |
| Locus 3 | miRNA-219: 3'-TCTTAACGCAAACCTGTTAGT-5' |
| Lnc-XIST: 5'-ATCATTACATTTGGACAATCA-3' | |
| Locus 4 | miRNA-219: 3'-TCTTAACGCAAACCTGTTAGT-5' |
| Lnc-XIST: 5'-ACGCTTACGTTTGGACAATCA-3' |
A luciferase reporter assay was used to determine whether lncRNA-XIST might function as a competitive endogenous RNA (ceRNA) for miR-219 (Figure 2B). Group A was the control group, and group B was transfected with only the wild-type reporter construct lncRNA-XIST(WT), so the fluorescence intensity of this group was high in the absence of the miR-219 mimic. Group C showed that the miR-219 mimic inhibited the fluorescence intensity of the wild-type reporter construct lncRNA-XIST(WT), suggesting that there was a correlation between lncRNA-XIST and miR-219.
The results of the RNA pull-down assay also showed that compared with the Bio-miR-219-inhibitor control, Bio-miR-219 expression was higher in HepG2/5-FU or HepG2 cells (P<0.05) (Table 4), while it was not markedly altered in the Bio-miR-219 (Mut) group (P>0.05), suggested that there was a direct relationship between lncRNA-XIST and miR-219 (Figure 2C).
Table 4
| Probe | Sequence |
|---|---|
| Bio-miR-inhibitor NC | 5'-UUCUCCGAACGUGUCACGUTT-3' |
| Bio-miR-219 | 5'-UGAUUGUCCAAACGCAAUUCUTT-3' |
| Bio-miR-219 (Mut) | 5'-UCUGGCCCCAAACGCAAUUCUTT-3' |
NC, negative control; Mut, mutation.
The lncRNA-XIST/miR-219/SMC4 axis plays an important role in the proliferation, invasion, and migration of HepG2/5-FU cells
To further investigate the relationship between lncRNA -XIST, miR-219, and SMC4, quantitative PCR analysis was performed to detect the expression of miR-219 and lncRNA-XIST. The results demonstrated that lncRNA-XIST expression was significantly upregulated and miR-219 expression was significantly downregulated in HepG2/5-FU cells compared to that in HepG2 cells (Figure 3A). Secondly, effective intervention fragments of lncRNA-XIST were constructed (target sequence 1: 5'-GCTGACTACCTGAG ATTT AAG-3'; target sequence 2: 5'-CTC TTGAACAGTTAATTTGC-3') (Table 5). LncRNA- XIST interference and miR-219 inhibitor/mimic fragments were cloned and stably transfected into the cell lines and compared with the controls. RT-PCR was performed, and the results showed that after lncRNA-XIST knockdown, the expression of miR-219 increased, resulting in the downregulation of SMC4 expression; after miR-219 knockdown, lncRNA-XIST/SMC4 would increase with the decrease of miR-219 (Figure 3B).
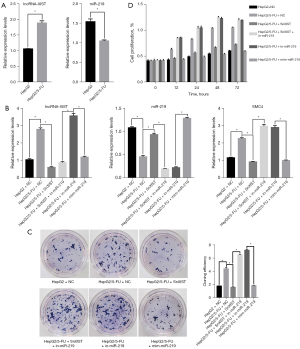
Table 5
| siRNA | Sense | Antisense |
|---|---|---|
| siRNA-XIST-1 | GCUGACUACCUGAGAUUUATT | UAAAUCUCAGGUAGUCAGCTT |
| siRNA-XIST-2 | CUCUUCAACAGUUAAUUUGTT | CAAAUUAACUGUUGAAGAGTT |
| Control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
To explore the potential role of the lncRNA-XIST/miR-219/SMC4 axis in HepG2/5-FU cells, cellular proliferation was evaluated using colony formation and MTT assays (Figure 3C,3D). To further study the invasion and migration ability of the cells, wound healing and invasion assays were performed (Figure 4A,4B), and flow cytometry assays were also carried out to evaluate cell apoptosis (Figure 4C).
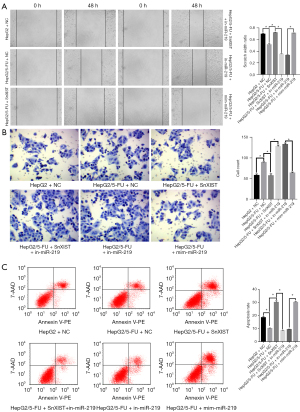
In the above experiments, groups A1 and A2 were the control groups. Group B showed that the knockdown of lncRNA-XIST resulted in inhibition of proliferation and downregulation of invasion compared with the A2 group. Therefore, the level of autophagy in this group would also be down-regulated and apoptosis increase. Group C showed that the knockdown of lncRNA-XIST resulted in the upregulation of miR-219 expression, which ultimately resulted in the inhibition of cell proliferation and invasion. The addition of exogenous miR-219 inhibitors reduced the expression of miR-219. The interaction between small nuclear RNA XIST (snRNA-XIST) and miR-219 could be offset. Therefore, we concluded that lncRNA-XIST promoted tumor drug resistance and growth by inhibiting miR-219. Group D showed that inhibition of miR-219 alone could promote cell proliferation and invasion. Compared with group C, group D had stronger cell proliferation and cell invasion ability because group C had snRNA-XIST, which partially offset the cancer-promoting effect of miR-219 inhibitors. In contrast to group D, group E showed that the overexpression of miR-219 significantly inhibited cell proliferation and invasion. The effect observed in this group was similar to that observed in group B because the knockdown of lncRNA-XIST resulted in an increase in miR-219 expression. However, statistical analysis demonstrated a significant difference in the association between the experimental groups compared with the normal control groups (P<0.05). Therefore, we conclude that the lncRNA-XIST/miR-219/SMC4 axis plays an important role in the proliferation, invasion, and migration of HepG2/5-FU cells.
To further investigate how the miR-219/SMC4 pathway affects autophagy in HCC cells, we found that JAK2/Stat3 promotes autophagy in glioma by positively regulating the adenosine 5'-monophosphate (AMP)-activated protein kinase/mechanistic target of rapamycin (AMPK/mTOR) pathway after reviewing the relevant literature (22). Therefore, we speculate that miR-219/SMC4 may be involved in the autophagy of HCC cells via the AMPK/mTOR pathway and affects the resistance to chemotherapy agents. It is well known that AMPK is an important energy-regulating molecule that promotes autophagy through autophosphorylation, while as an inhibitor of autophagy, mTORC1 can inhibit autophagy by activating protein synthesis (23). We substantiated our hypothesis using western blotting (Figure 5).
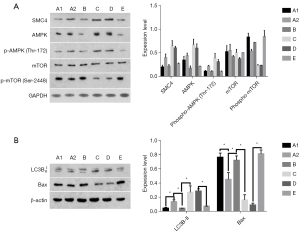
5-FU was added to the following groups and incubated for 24 hours. Group A1 and A2 were the control groups. In group A2, the level of autophagy in this group was markedly increased [phosphorylated adenosine 5'-monophosphate (AMP)-activated protein kinase (p-AMPK), LC3B-II up-regulated, p-mTOR down-regulated] and apoptosis was decreased [BCL2-associated X (Bax) down-regulated] compare to the group A1.
In group B, we knocked down the expression of the lncRNA-XIST gene and induced the up-regulation of miR219 expression in parallel with drug exposure in the drug resistant cells. Compared with the A2 group, the level of autophagy in this group was markedly decreased (p-AMPK, LC3B-II down-regulated, p-mTOR up-regulated) and apoptosis was increased (Bax up-regulated). In other words, snRNA-XIST inhibited autophagy and restored the drug sensitivity of drug-resistant cells. This indicated that up-regulation of lncRNA-XIST expression was the major factor of drug resistance, and suggested that up-regulation of miR-219 could also restore the drug sensitivity of drug-resistant cells.
In group C, the results showed that the level of autophagy in this group was markedly increased (p-AMPK, LC3B-II up-regulated, p-mTOR down-regulated) and apoptosis was decreased (Bax down-regulated) compared to the group B. The results also indicated that lncRNA-XIST increased levels of autophagy and drug resistance in HCC cells by inhibiting miR-219. Group D was set as a control for group C, the level of autophagy in this group was changed unobviously (p-AMPK, LC3B-II, p-mTOR unchanged) compared to the group C. The results indicated that the inhibition of miR-219 could improve autophagy levels and drug resistance significantly in HCC cells. Group E was set as a control for group D and B, the effect of this group was basically similar to that of group B, the level of autophagy was changed unobviously (p-AMPK, LC3B-II, p-mTOR unchanged) compared to the group B. That indicated that the expression of miR-219 was enhanced by transfection of miR-219 mimics in drug-resistant cells, which could significantly inhibit autophagy and induce apoptosis. The results also showed that the up-regulated expression of miR-219 could restore the drug sensitivity, while apoptosis was increased in this group.
Our experiments substantiate the hypothesis that miR-219/SMC4 is involved in the autophagy process of HCC cells through the AMPK/mTOR pathway and enhances the drug resistance of liver cancer cells.
Discussion
Autophagy is an evolutionarily conserved, lysosome-dependent, catabolic process in which cytoplasmic components such as damaged organelles, protein aggregates, and lipid droplets, are degraded and their components recycled (24). In recent years, a study has demonstrated that autophagy plays an important role in all stages of tumor development, and the relationship between autophagy and tumorigenesis may be two-fold (25). Autophagy has been found to play a dual role in the occurrence and development of HCC (26). It acts as a tumor suppressor to inhibit tumorigenesis, likely before normal cells are transformed into cancer cells, and also promotes cancer progression and malignancy in already transformed cancer cells (27). During the advanced stage, autophagy can promote the metabolism of hepatoma cells, inhibit apoptosis, and increase the resistance to chemotherapy drugs (28). It is generally accepted that tumor cells utilize autophagy to overcome the stresses caused by chemotherapeutic agents, radiation, and molecular-targeted agents. Hence, the changes in their activation may be useful to enhance chemotherapeutic drug sensitivity in clinical research studies. Herein, we focused on the function of SMC4 in enhancing the chemoresistance of hepatoma cells by promoting autophagy.
SMC4 encodes a “structural maintenance of chromosomes” protein that is a member of the SMC family. This adenosine triphosphatase (ATPase) family maintains the stability of chromosomal structure and participates in eukaryotic cell mitosis (29,30). SMC4 plays important roles in chromosome condensation and mitosis and is required for normal S phase progression, indicating a previously unrecognized role of SMC4 in the synchronous progression from the G1 to S phase (31,32). Consistent with our previous study, we found that SMC4 may contribute to the development of HCC (18). To further study the mechanism underlying the role of SMC4 in the autophagy of hepatoma cells, we established a drug-resistant hepatoma cell line, HepG2/5-FU, and found that the expression level of SMC4 was significantly increased. Immunofluorescence was also used to detect the level of autophagy, and the results indicated that the level of autophagy was also significantly increased compared to that in the HepG2 cells. Taken together, these results confirmed that SMC4 might play an important role in the autophagy of hepatoma cells. However, the specific role of SMC4 in autophagy still requires further illustration.
In further experiments of the upstream regulation of miR-219/SMC4, the expressions of two typical lncRNAs (lncRNA-XIST and lncRNA-TUG1) were found to be upregulated by microarray. Through these experiments, we confirmed that lncRNA-XIST acted as an intrinsic miRNA sponge or regulatory factor of miR-219. Previous studies have shown that lncRNA-XIST plays important roles in several diseases, is involved in the progression of many types of cancer, and affects disease prognosis (33-35). Thus, targeting lncRNAs may be a promising strategy for patients with chemoresistance (36-38). However, to date, no study has reported a relationship between lncRNA-XIST and miR-219/SMC4 and its effect on autophagy in hepatoma cells. In this study, we found that lncRNA-XIST expression was upregulated in HepG2/5-FU cells, and when lncRNA-XIST expression was decreased, miR-219 could effectively bind to SMC4 and reduce the expression of SMC4. In turn, the autophagy level of hepatoma cells decreased, and the drug resistance of hepatoma cells decreased. To investigate the mechanism of miR-219/SMC4-induced autophagy in HCC cells, we speculated that miR-219/SMC4 may be involved in HCC cell autophagy through the AMPK/mTOR pathway and affected its resistance to chemotherapy drugs by reviewing the literature. Intracellular ATP was found to be significantly reduced in HCC cells when they were starved. Aiming to provide energy to HCC cells, tumor cells eventually activate HCC cells for autophagy via AMPK/mTOR (39-41). The AMPK/mTOR signaling pathway plays an important role in autophagy of HCC cells (41).
Through our experiments, we confirmed our hypothesis that miR-219/SMC4 was involved in the autophagic process of HCC cells through the regulation of AMPK/mTOR and enhanced the drug resistance of HCC cells.
Conclusions
In conclusion, our data indicated that SMC4 could promote autophagy and increase the drug resistance of HCC cells. Moreover, lncRNA-XIST may competitively bind to miR-219 by acting as a miRNA sponge, thereby preventing miR-219 from effectively reducing the expression of SMC4 and facilitating autophagy and drug resistance in HCC cells via the AMPK/mTOR pathway. And this study will further elucidate the role of autophagy in drug resistance of HCC to enhance drug sensitivity. It is of great significance to improve the sensitivity of hepatocellular carcinoma cells to chemotherapy drugs and improve the therapeutic effect of patients. Overall, this study suggests that SMC4 may act as a potential marker of a poor response of HCC to chemotherapy and a novel therapeutic target for HCC chemotherapy.
Acknowledgments
Funding: This work was financially supported by the Chongqing Natural Science Foundation (No. cstc2020jcyj-msxmX0122).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3623/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3623/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3623/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Ringelhan M, Pfister D, O'Connor T, et al. The immunology of hepatocellular carcinoma. Nat Immunol 2018;19:222-32. [Crossref] [PubMed]
- Globocan. International agency for research on cancer. Published 2020. Cited 2021 Feb 15. Available online: http://gco.iarc.fr/today/home
- Wong RJ, Ahmed AHepatocellular Carcinoma Research Committee of the Chronic Liver Disease Foundation. Understanding Gaps in the Hepatocellular Carcinoma Cascade of Care: Opportunities to Improve Hepatocellular Carcinoma Outcomes. J Clin Gastroenterol 2020;54:850-6. [Crossref] [PubMed]
- Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol 2021;15:1295-307. [Crossref] [PubMed]
- Global burden of disease (GBD) compare. Institute for health metrics and evaluation. Published October 2019. [Cited 2021 Feb 14]. Available online: http://vizhub.healthdata.org/gbd-compare
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3:524-48. [Crossref] [PubMed]
- Xu J. Trends in Liver Cancer Mortality Among Adults Aged 25 and Over in the United States, 2000-2016. NCHS Data Brief 2018;1-8.
- Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723-50. [Crossref] [PubMed]
- Chen Z, Xie H, Hu M, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res 2020;10:2993-3036.
- Li X, Ramadori P, Pfister D, et al. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer 2021;21:541-57. [Crossref] [PubMed]
- Hou Z, Liu J, Jin Z, et al. Use of chemotherapy to treat hepatocellular carcinoma. Biosci Trends 2022;16:31-45. [Crossref] [PubMed]
- Xu XF, Yang XK, Song Y, et al. Dysregulation of non-coding RNAs mediates cisplatin resistance in hepatocellular carcinoma and therapeutic strategies. Pharmacol Res 2022;176:105906. [Crossref] [PubMed]
- Xue J, Ni H, Wang F, et al. Advances in locoregional therapy for hepatocellular carcinoma combined with immunotherapy and targeted therapy. J Interv Med 2021;4:105-13. [Crossref] [PubMed]
- Sangro B, Sarobe P, Hervás-Stubbs S, et al. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2021;18:525-43. [Crossref] [PubMed]
- Sheng J, Qin H, Zhang K, et al. Targeting autophagy in chemotherapy-resistant of hepatocellular carcinoma. Am J Cancer Res. 2018;8:354-65.
- Zhou B, Yuan T, Liu M, et al. Overexpression of the structural maintenance of chromosome 4 protein is associated with tumor de-differentiation, advanced stage and vascular invasion of primary liver cancer. Oncol Rep 2012;28:1263-8. [Crossref] [PubMed]
- Zhou B, Chen H, Wei D, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J Exp Clin Cancer Res 2014;33:55. [Crossref] [PubMed]
- Janji B, Berchem G, Chouaib S. Targeting Autophagy in the Tumor Microenvironment: New Challenges and Opportunities for Regulating Tumor Immunity. Front Immunol 2018;9:887. [Crossref] [PubMed]
- Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol 2018;24:4643-51. [Crossref] [PubMed]
- Fu XT, Shi YH, Zhou J, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett 2018;412:108-17. [Crossref] [PubMed]
- Yuan G, Yan SF, Xue H, et al. Cucurbitacin I induces protective autophagy in glioblastoma in vitro and in vivo. J Biol Chem 2014;289:10607-19. [Crossref] [PubMed]
- Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett 2010;584:1287-95. [Crossref] [PubMed]
- Kitada M, Koya D. Autophagy in metabolic disease and ageing. Nat Rev Endocrinol 2021;17:647-61. [Crossref] [PubMed]
- Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J 2021;40:e108863. [Crossref] [PubMed]
- Yu S, Wang Y, Jing L, et al. Autophagy in the "inflammation-carcinogenesis" pathway of liver and HCC immunotherapy. Cancer Lett 2017;411:82-9. [Crossref] [PubMed]
- Chao X, Qian H, Wang S, et al. Autophagy and liver cancer. Clin Mol Hepatol 2020;26:606-17. [Crossref] [PubMed]
- Yang S, Wang X, Contino G, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011;25:717-29. [Crossref] [PubMed]
- Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 2005;19:1269-87. [Crossref] [PubMed]
- Nishiwaki T, Daigo Y, Kawasoe T, et al. Isolation and characterization of a human cDNA homologous to the Xenopus laevis XCAP-C gene belonging to the structural maintenance of chromosomes (SMC) family. J Hum Genet 1999;44:197-202. [Crossref] [PubMed]
- Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 2000;149:811-24. [Crossref] [PubMed]
- Yu L, Peña Castillo L, Mnaimneh S, et al. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell 2006;17:4736-47. [Crossref] [PubMed]
- Chen YK, Yen Y. The Ambivalent Role of lncRNA Xist in Carcinogenesis. Stem Cell Rev Rep 2019;15:314-23. [Crossref] [PubMed]
- Zhu J, Kong F, Xing L, et al. Prognostic and clinicopathological value of long noncoding RNA XIST in cancer. Clin Chim Acta 2018;479:43-7. [Crossref] [PubMed]
- Wang W, Min L, Qiu X, et al. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front Cell Dev Biol 2021;9:645647. [Crossref] [PubMed]
- Zhang X, Xie K, Zhou H, et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol Cancer 2020;19:47. [Crossref] [PubMed]
- Yuan L, Xu ZY, Ruan SM, et al. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol Cancer 2020;19:96. [Crossref] [PubMed]
- Wang Y, Fang Z, Hong M, et al. Long-noncoding RNAs (lncRNAs) in drug metabolism and disposition, implications in cancer chemo-resistance. Acta Pharm Sin B 2020;10:105-12. [Crossref] [PubMed]
- Ashworth RE, Wu J. Mammalian target of rapamycin inhibition in hepatocellular carcinoma. World J Hepatol 2014;6:776-82. [Crossref] [PubMed]
- Chiarini F, Evangelisti C, McCubrey JA, et al. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci 2015;36:124-35. [Crossref] [PubMed]
- Cui J, Shen HM, Lim LHK. The Role of Autophagy in Liver Cancer: Crosstalk in Signaling Pathways and Potential Therapeutic Targets. Pharmaceuticals (Basel) 2020;13:432. [Crossref] [PubMed]


