Mending a broken heart—targeting cardiomyocyte regeneration: a literature review
Introduction
Massive or progressive cardiomyocyte loss constitutes the underlying pathogenesis of heart failure, which leads to the increased mortality of a variety of cardiovascular diseases (1). These devastating conditions result in a comparable or even worse prognosis than do those of common malignancies (2,3). Unfortunately, the adult mammalian heart has extremely limited regenerative potential compared to that of lower vertebrates, who possess a profound ability to regenerate even in the adult stage (4,5). The idea of replenishing or regenerating injured myocardium has attracted broad interest for decades. After extensive discussion and heated debate, the field has achieved consensus regarding some fundamental principles. In this review, we will focus on the key biomedical findings and underlying principles of cardiomyocyte regeneration.
This review aims to (I) provide an overview of the current literature involved in the controversies regarding the origins of newly formed cardiomyocytes and potential barriers to heart regeneration; (II) summarize the animals, models, and the key methods applied in the study of heart regeneration; and (III) address potential strategies to boost heart regeneration. This review makes a novel contribution to the current literature by detailing the barriers and potential strategies to heart regeneration and offers prospects for future research. We present the following article in accordance with the Narrative Review reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2649/rc).
Methods
We searched the PubMed and Web of Science databases for original research and review articles published in English and Chinese languages between January 1990 and March 2022. The following search terms were employed in different combinations: “heart regeneration” or “cardiac regeneration”, “dedifferentiation”, “cell proliferation”, “nuclear reprogramming”, “cardiac injury”, and “cardiac repair”. Articles citing related studies and citations in relevant articles were also examined as potential sources of information. The database resources are summarized in Table 1 and Table S1.
Table 1
| Items | Specification |
|---|---|
| Date of search | March 25 to March 30, 2022 |
| Databases and other sources searched | PubMed and Web of Science databases |
| Search terms used | See Table S1 for details |
| Timeframe | From January 1990 to March 2022 |
| Inclusion and exclusion criteria | Inclusion criteria: Original articles and review articles in English and Chinese languages |
| Exclusion criteria: (I) publications with duplications or studies with overlapping data from the same author; (II) abstracts, case reports, proceedings, and meta-analyses; and (III) incomplete outcome data | |
| Selection process | Two reviewers included and validated the studies independently. Inconsistencies in the opinions of the two reviewers were resolved by negotiation and discussion by all authors |
Spontaneous mammalian cardiomyocyte regeneration
In the mammalian heart, cardiomyocytes proliferate during the embryonic and early postnatal stages. At the time of birth, the neonatal human heart mainly consists of mononuclear cardiomyocytes and binuclear cardiomyocytes (~30%). Soon after, a majority of cardiomyocytes exit the cell cycle and lose regenerative capacity but still undergo whole genome duplication without cytokinesis, leading to binucleation (6). The ratio of mononucleated and binucleated cardiomyocytes remains nearly consistent after birth. Polyploidization increases significantly in human cardiomyocytes during the postnatal stage, with the proportion reaching over 60% for mononucleated tetraploids in adult cardiomyocytes (4). Terminal differentiation of perinatal cardiomyocytes inhibits cytokinesis, thereby resulting in binucleation and the limitation of regenerative repair after injury. The newly generated cardiomyocytes dramatically decrease within the first few days after birth (7,8). The adult human heart is considered to be incapable of further cell division but grows with hypertrophy. However, a growing body of evidence suggests that cardiomyocytes self-renew at a very limited capacity.
In the adult heart, the extent of cardiomyocyte exchange varies depending on the pathophysiological condition (9). In rodent models, the reported turnover rates of adult cardiomyocytes range from 0 to 4.0% annually (7,8,10-12), which increases to a marginally higher level after injury (13). Phosphorylated histone H3 has been identified in the human adult heart up to the age of ~20 years by histological analysis, which implies the existence of mitosis (5). Nevertheless, the basic assumption depends on the presence of cell division or death markers, which may not accurately represent actual cell division or death. Recently, a new strategy based on carbon-14 dating has been applied to study cell generation in experimental animals, revealing for the first time that the renewal rate of cardiomyocytes decreases gradually from 1% to 0.45% from youth to older age. Fewer than 50% of cardiomyocytes undergo turnover over a whole lifespan (4). Using the same approach, Bergmann et al. (14) reported that the renewal rate of cardiomyocytes peaks in early childhood, which is similar throughout both of the ventricles regardless of ventricular configuration and hemodynamic workload. However, despite the cardiomyocytes possessing this apparent renewal capacity, effective proliferation is activated in myocardial infarction (MI). Senyo et al. (11) found that only 3% of pre-existing cardiomyocytes in the infarct zone undergo polyploidization without cytokinesis.
Lessons from lower vertebrate cardiomyocyte regeneration
Certain lower vertebrates, such as fish and amphibians, maintain strong cardiac regeneration throughout their entire lifespan, and the neonatal mammalian heart also possesses the regenerative potential of heart tissue after injury that involves the widespread dedifferentiation and proliferation of cardiomyocytes (15,16). After apical resection in 1-day-old mice, the neonatal heart exhibits robust regenerative potential without hypertrophy or obvious fibrosis. Although myocyte necrosis and collagen deposition can cause some scarring, the tissue is almost completely repaired within 3 weeks with minimal fibrosis in both the MI and the ventricular resection models, and the cardiac function returns to normal 9 months after injury (15).
Controversies regarding the origins of newly formed cardiomyocytes
The origin of newly formed cardiomyocytes after injury is controversial. Among the potential origins are resident cardiac progenitor cells (CPCs) (17-20). Some data support the notion that noncardiomyocyte progenitor cells hardly contribute to new cardiomyocytes in the infarcted zone (21,22). Additionally, genetics approaches have further confirmed that the adult cardiac stem cell population, including c-kit+ cells, seldom gives rise to cardiomyocytes (23). Instead, this population may enhance cardiomyocyte survival and neovascularization via paracrine signaling to improve cardiac function after CPC transplantation (19). Importantly, genetic fate mapping has shown that most newly formed cardiomyocytes originate from pre-existing cardiomyocytes and not CPCs (11,24). Based on these findings, stimulating existing cardiomyocytes could be a potential strategy to regenerating the injured myocardium.
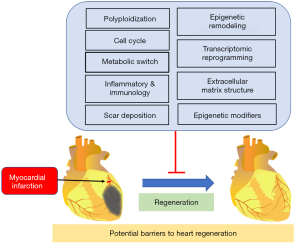
Potential barriers to heart regeneration
As summarized in Figure 1, recent studies have suggested that there are several potential barriers to heart regeneration, including metabolic switch, a considerable increase in multinucleated and polyploid cardiomyocytes, and epigenome and transcriptome alteration (8,14,25).
Polyploidization and the cell cycle
In postnatal mammals, the polyploidization of heart muscle occurs naturally. Cardiomyocytes experience additional DNA synthesis without cytokinesis shortly after birth, resulting in a large number of binucleated and tetraploid cardiomyocytes. The disappearance of regenerative capacity manifests as polyploidization and the loss of cytokinesis (26), as evidenced in murine neonates (15). Moreover, the majority of murine cardiomyocytes undergo hypertrophic growth shortly after birth to meet the increasing needs of circulatory support at the expense of regenerative ability. The rare proliferative cardiomyocyte subpopulation exhibits a hypoxic gene signature, which may determine the cell transformation toward either cytokinesis or hypertrophic growth (27,28). Furthermore, a coronary artery ligation model found that cardiomyocyte proliferation is positively correlated with the number of preinjury mononuclear diploid cardiomyocytes (29).
In the adult cardiomyocytes of zebrafish and newts, the centrosome integrity is maintained but is lost in mammalian neonatal cardiomyocytes. The loss of centrosome integrity leads to the failure of ciliogenesis. Meanwhile, centrosome disassembly is coupled with cell cycle exit, suggesting that in mammalian cardiomyocytes, the loss of centrosome integrity is the basis of the postmitotic state during development (30). Moreover, mature cardiomyocytes without proliferative potential adopt a contractile and energetically active state (31,32). From this perspective, the loss of regenerative potential in the adult mammalian heart may be an adaption for higher contractility and increased metabolic activity to meet circulatory demands (33).
Metabolic switch
Immediately after birth, the heart experiences a transition from a relatively hypoxic to a normoxic environment. To accommodate this oxygen change, the cardiac transcriptome undergoes extensive remodeling, which involves the shutdown of cell cycle genes and induction of metabolic genes, thereby enhancing oxidative phosphorylation (34). Recent data have shown that oxidative phosphorylation may exhibit a direct role in cell cycle exit in postnatal cardiomyocytes (35). Consistently, cardiomyocyte maturation is accompanied by increased levels of free fatty acids and enhanced fatty acid oxidation, which is in contrast to the glycolysis-dependent process of immature cardiomyocytes (36). Repression of fatty acid β-oxidation metabolism in the infant mouse heart prolongs the cardiomyocyte cell cycle but delays hypertrophic growth and maturation (37,38).
Transcriptomic reprogramming
Transcriptomic reprogramming is tuned to facilitate the transition from neonatal stages to adulthood. Intense differences between neonatal and adult myocyte transcription have been observed (6,276 differentially expressed transcripts) (34). Transcriptional networks, which control cell cycle transition and metabolism, are differentially expressed in both neonatal and adult myocytes (39). Moreover, the genome architecture is reprogrammed accordingly, as it is condensed in the regions covering the cell cycle genes and becomes open at the promoter regions of metabolism and contractility with maturation (34). DNA damage related to perinatal oxidative stress and higher oxygen levels may result in cell cycle exit in cardiomyocytes (35). Meanwhile, the cessation of cytokinesis may occur due to excessive oxidative stress (40).
Epigenetic remodeling
Epigenetic regulation plays a significant role in the regulation of the transcriptional state during development and cell transformation. Chromatic architecture is generally regulated by histone modifications and DNA methylation (41,42). During postnatal maturation, the lack of chromatin accessibility can limit cardiac regenerative potential after birth. Hence, chromatin modification may need to be overcome to facilitate cell cycle re-entry in adults (34).
The patterns of DNA methylation in cardiomyocytes also exhibit dynamic signatures during cardiac development and maturation. Cardiac DNA methylation patterns have been reported to have extensive alterations during maturation, which have been identified in a recent genomewide study (43). Transcription factor (TF) binding sites are highly methylated in embryonic stem cells as compared to the demethylation in differentiated cardiomyocytes. Furthermore, the accessibility of genes encoding fetal isoforms of sarcomeric proteins becomes closed, and repressive histone marks are expressed during cardiac maturation. In contrast, genes regulating calcium handling and contraction, including Atp2a2, Myh6, Tnnt2, and Tnni3, are transcriptionally activated through demethylation from the embryonic to adult stages. The DNA becomes demethylated and obtains histone activation marks, such as H3K27ac, H3K4me3, and H3K4me1. Moreover, a certain number of developmental signaling pathway-related genes are hypermethylated during neonatal heart maturation with transcriptional repression. The removal of hypermethylated land markers inhibits binucleation and promotes cardiomyocyte proliferation in vivo (44).
There are several key epigenetic modifiers, such as long noncoding RNAs (lncRNAs), which regulate transcription and may play roles in cardiac repair and regeneration (45). For example, Fendrr is necessary for embryonic heart development and cardiac proliferation (46). Myocardial hypoplasia and ventricular thinning, as well as the reduced mitosis of cardiomyocytes, can be caused by the homozygous deletion of Fendrr. In neonatal murine cardiomyocytes, the inhibition of Bvht upregulates fetal cardiac genes (47). This will be an important topic for future investigations that explore the roles of lncRNAs in cardiac regeneration.
Histone acetyltransferases and histone deacetylases (HDACs) participate in cardiomyocyte proliferation by regulating chromatin accessibility. p300 interacts with critical cardiac TF and then directly acetylates them to enhance DNA binding (48-50). Genetic knockout of p300 leads to proliferation defects, reduced expression of cardiac structural genes, and cardiac structural flaws, such as impaired trabeculation (51). HDACs can promote chromatin condensation by removing acetylation marks and repressing transcription. The deletion of Hdac1 and Hdac2 in cardiomyocytes is related to neonatal lethality and severe cardiac dysfunction (52). Mice without cardiac Hdac3 will die from severe cardiac hypertrophy in the adult stage (53). Conversely, cardiac-specific overexpression of Hdac3 causes enlargement of the heart via excessive cardiomyocyte proliferation (54). However, the precise mechanisms related to histone acetylation and heart regeneration are still undefined.
Inflammatory and immunology
Along with intensive research of diverse species, a popular idea is that immune system evolution in mammals is associated with the loss of regenerative capacity when compared to lower vertebrates (55). Moreover, the proinflammatory function in neonates is impaired. The immature adaptive immune system may provide an immunological environment that is prone to regeneration (56).
To facilitate tissue regeneration after injury, neonate hearts do not mount a robust fibrotic response, but rather a more angiogenic one; meanwhile, adult hearts exhibit the opposing tendency. However, our understanding of the molecular basis underlying the divergent outcomes of the inflammatory or immune response remains limited. Macrophages and their polarization states have been regarded as a central regulator of the tissue healing process. A previous study showed that macrophage depletion in neonates does not influence cardiomyocyte proliferation with reduced cardiac function and angiogenesis after injury (57). The authors further compared the immunophenotyping and gene expression profiling of cardiac macrophages from regenerating and non-regenerating hearts following MI. They found that macrophages from neonatal hearts exhibit unique polarization with no clear bias toward M1 or M2, while macrophages from post-neonatal hearts have dramatically upregulated levels of M2. In addition, macrophages from tissue-resident or circulating monocytes play different roles in the neonatal heart. Resident macrophages contribute to cardiomyocyte regeneration, whereas monocyte-derived macrophages boost the inflammatory response and mediate the lack of reparative activities after injury. The activity of T cells, which belong to the adaptive immune system, also plays critical roles during tissue repair and regeneration (58).
Animals used in the study of heart regeneration
An important question is how to study heart regeneration. Current knowledge regarding heart regeneration is emerging from diverse research avenues that each use distinct species and models. With the development of novel molecular and genetic tools, most barriers to obtaining insights have been overcome. The following section summarizes and evaluates the tools used by the most popular systems.
Amphibians
Urodele amphibians, including newts and axolotls, possess the capacity for heart regeneration. Newts undergo complete regeneration 60 days after resection or mechanical crushing of the lateral ventricles (59,60). An abundant downregulation of sarcomere proteins can be caused by cardiac injury, indicating that the dedifferentiation and proliferation of existing cardiomyocytes are the main sources of cardiomyocyte proliferation. However, these animals’ long breeding cycles and extensive genomes makes their study difficult.
Zebrafish
The zebrafish is an ideal model system for molecular genetics. Zebrafish hearts can regenerate after injury induced by apical resection, cryoinjury, cardiomyocyte ablation, or hypoxia reperfusion (61-63). Zebrafish cardiomyocytes adjacent to the injury region experience a fetal transition, involving closed contractile gene transcription, and re-enter the cell cycle. Although collagen deposition can be detected during the regeneration process, permanent scarring is not obvious (26,63).
Neonatal mice
Neonatal mice have sound heart regeneration, and little or no residual scarring can be detected at the injury site after apical resection or surgical coronary artery ligation. The regenerative capacity is restricted to the first 7 days after birth (15,24). However, a relatively large injury size induced by a severe transmural cryoinjury leads to be very limited regeneration (64). Neonatal mice are an ideal model for studying heart regeneration and have been explored in depth using multiple advanced genetic tools.
Neonatal pigs
Recently, neonatal porcine hearts have been reported to possess the ability to regenerate. New cardiac muscle has been detected in 2-day-old pigs after permanent left anterior descending artery ligation, with functional recovery and without significant fibrosis. In contrast, abundant fibrosis, thinned myocardium, and reduced cardiac function have been observed in 14-day-old pigs with the same injury (65). Compared to other animal models, such as those involving zebrafish and mice, the neonatal pig model of regeneration is more anatomically and physiologically similar to the human heart. However, the expenses involved in neonatal pig studies are considerably higher, and the tools for genetic manipulation are limited.
Models used to study heart regeneration
Myocardium injury models
Several myocardium injury models have been routinely used. Apical amputation achieves muscle deletion by direct tissue removal in the apex, causing cell death at the injury site (15). The MI model is routinely created by left anterior descending coronary artery ligation, providing an outstanding ischemic injury model for revealing the molecular and cellular pathways in cardiac regeneration (66). Finally, the cryoinjury model was first developed in 2011 (61,67) and can better mimic MI and the diverse pathological changes involved, such as cardiomyocyte proliferation, massive death, inflammatory response, and reversible collagen deposition.
Genetic ablation of the epicardium
Specific cell types can be depleted through targeted expression of bacterial nitroreductase by transferring a nontoxic substrate (metronidazole) to a cytotoxin. Genetic ablation of the epicardium was firstly created using tcf21 regulatory sequences without direct myocardial damage (68). After ablation of over 90% of the epicardial cells with metronidazole treatment, adult zebrafish heart regeneration is substantially compromised, implying that the epicardium plays a critical role in cell communication to initiate cardiomyocyte proliferation.
Key methods applied to study heart regeneration
Genetic fate mapping
Genetic fate mapping is useful for tracing a targeted progenitor cell population by creating a permanent label. For this purpose, a tamoxifen-inducible Cre recombinase is inserted into a specific regulatory cassette, sometimes with a fluorescent reporter. The fluorescent reporter is normally controlled by loxP-flanked stop sequences (69,70). Comprehensive fate mapping has revealed that c-Kit-expressing cells in injured adult mouse hearts possess a degree of vascular endothelial potential but limited cardiomyocyte potential (71).
Carbon-14 dating
Age class is the core mathematical model used to describe cells. The bomb curve is used to obtain the carbon-14 concentration in the cell population. Bergmann et al. (4) was the first to quantify human cardiomyocyte replication by integrating carbon-14 into DNA during Cold War nuclear testing. However, an increase in polyploidization cannot be ruled out, which may be a confounding factor in study results (14).
Single-cell omics
Emerging evidence has shown that cell network and communication play an important role in heart regeneration. Single-cell RNA-sequencing (RNA-seq) has the unprecedented ability to dissect cell interactions, identify cell subpopulations with regenerative potential, and describe cell state transition, which may help to answer the central question in this field (72-74). However, single-cell sequencing has some challenges, such as lowly expressed genes and the capture of any noncoding RNA information. A novel sequencing solution may expand the application of single-cell RNA-seq in the future (75).
Lineage tracing models
Lineage tracing was developed to track proliferating cells and can be used to mark several cells at a specific time point (76). Mosaic analysis with double markers (MADMs) is a powerful tool of lineage tracing for marking recombination events in both mitotic and postmitotic cells (77). MADMs enables in vivo analysis of gene function with simultaneous labeling and gene knockout at the single-cell level and has been widely applied in cardiac regeneration research (78,79). Another stochastic model for lineage tracing is the rainbow reporter system, which works through fluorophore-mediated cell labeling. The rainbow reporter system has been proven to be a useful lineage-tracing tool for investigating cardiomyocyte proliferation during normal development and in pathological conditions (80-83).
Quantitative histological analysis of cardiomyocyte proliferation
The quantitative histological method for assessing the cardiomyocyte proliferation associated with cardiomyocyte counting is a critical technique used in the study of cardiac proliferation. Nucleotide incorporation using 5-bromo-2'-deoxyuridine (BrdU) or 5-ethynyl-2-deoxyuridine (EdU) is widely acknowledged as a reliable measure of cell cycle re-entry and proliferation (84-86). Through the colocalization of mitotic marker pHH3 or S-phase marker proliferating cell nuclear antigen (PCNA) with the cardiomyocyte marker myosin heavy chain, proliferated cardiomyocytes with incorporated additional nucleotides can be visualized and quantified via immunofluorescent microscopy (87).
Strategies to boost heart regeneration
Animal studies involving increased cardiomyocyte regeneration capacity have provided new insights into strategies that boost cardiac regeneration (as summarized in Figure 2).
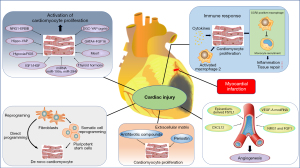
Activation of resident cardiomyocytes
Recent advances in the field now recognize that CPCs only possess limited regenerative potential, and the activation of cardiomyocytes could be a target for improving heart regeneration. Initially, cell cycle regulation was applied to study reactivated cardiomyocyte proliferation (88-91). The inhibition of Meis1 results in an extended regenerative window in neonates and a reactivation of the cell cycle in adults (92). Hand2, another developmental gene, induces cardiomyocyte proliferation during regeneration by triggering a certain number of DNA replications, little cytokinesis, and consequently, very limited new cardiomyocytes (93). Neuregulin1 (NRG1) is an epidermal growth factor (EGF)-like growth factor binding ErbB4 and ErbB2 receptors. It has been shown that NRG1 plays important roles in maintaining cardiac homeostasis in adult mammals (94,95). The delivery of fibroblast growth factor 1 (FGF1) along with the inhibition of the p38 mitogen-activated protein kinase (MAPK) pathway promotes the repair of the injured myocardium (96). Hippo signaling regulates cardiomyocyte proliferation to control organ size via Yes-associated protein/transcriptional coactivator with PDZ-binding motif (YAP/TAZ) signaling. For instance, YAP overexpression in mouse cardiomyocytes provokes their proliferation and induces cardiomegaly (97). Moreover, the Hippo pathway could be a viable target for regeneration, as regulation of this pathway protein in adult cardiomyocytes by genetic mutation or overexpression can enhance cardiomyocyte proliferation and reduce the injury-related scar size after MI (95). Agrin, a matrix glycoprotein, is necessary to induce the regeneration of neonatal mouse hearts, and the administration of agrin in adult mice promotes cardiac regeneration following MI injury (98,99).
MicroRNAs (miRNAs) such as miR-590 and miR-199a, which contribute to the promotion of cell cycle re-entry, can also be therapeutic targets for cardiomyocyte proliferation (100). Tao et al. (101) revealed that overexpression of the miR-302-367 cluster in adult cardiomyocytes is sufficient to regenerate the myocardium after MI injury and works via suppression of the Hippo pathway. Similarly, the overexpression of human miR-199a or miR-15 can stimulate cardiac repair in a pig MI model (24,102).
Induced pluripotent stem cell (iPSC)-induced cardiomyocyte reprogramming
With the progression of iPSC techniques, iPSC-induced cardiomyocytes could be an important source of cardiac repair. In addition, tissue scaffolds can support vascular cells with or without cardiomyocytes to create cardiac patches with contractility, better mimic cardiac complexity, and improve the survival and maturation of donor cells (103). At present, there are still several challenges to be overcome. The production scale is one such challenge, as the generation of tens of thousands of surviving cardiomyocytes is required for each patient. Another challenge is transient or sustained ventricular arrhythmias, which occur as a result of unsuccessful cell integration. Ventricular arrhythmias can cause cardiac arrest, whereas the implantation of internal cardioverter-defibrillator devices can alleviate arrhythmia-related deaths (104,105).
Direct reprogramming
Direct reprogramming, also known as trans-differentiation, is a process that converts somatic cells from one lineage to cardiomyocytes without transitioning through an intermediate pluripotent state. Cardiac fibroblasts have been the major source of converting induced cardiomyopathy (iCM) in vivo (106). Compared to iPSC reprogramming, direct reprogramming exhibits unique advantages in a faster and more efficient way while also enabling the conversion of cells in situ without pluripotent transitioning or ex vivo cell expansion (107). Meanwhile, epigenetic marks of the cell of origin, such as aging marks, are maintained.
In 2010, Ieda et al. (106) introduced the TFs Gata4, Mef2c, and Tbx5 into cardiac fibroblasts, which were reprogrammed into iCMs expressing contractile genes in vitro, and successfully transplanted these into infarcted murine hearts. Mouse cardiac fibroblast-derived iCMs in vitro are like neonatal cardiomyocytes, whereas iCMs obtained in vivo are like adult cardiomyocytes with an equivalent transcriptional signature and morphology that can electrically couple with endogenous cardiomyocytes (108,109). It is more difficult to convert human fibroblasts into iCMs. Recently, heart and neural crest derivatives expressed 2 (HAND2) has been listed among the factors that can improve the efficiency of reprogramming (110). Cocktails that efficiently convert human fibroblasts into iCMs also have been identified (111,112).
Profound changes in the epigenetic landscape occur in cellular reprogramming. Combinations of epigenetic modulators with specific miRNAs or signaling pathways inhibitors have demonstrated an ability to efficiently reprogram human fibroblasts to iCMs (107,113,114). Thus, extensive efforts have been exerted to identify potential epigenetic barriers in direct reprogramming (including ICR2, BAF60c, HELLS, etc.), although the mechanism remains unclear (114,115).
Adjuvant interventions
Promoting neovascularization and modulating the immune environment may further enhance local myocardial regeneration. From a practical perspective, small bioactive molecules or gene therapy may be a promising approach. Indeed, inducing vascular endothelial growth factor (VEGF) expression by employing modified RNA technology has been found to improve the regenerative response following injury via angiogenesis (116). However, more evidence is still needed for clinical application since the double-blind EUROINJECT-ONE and NORTHERN clinical trials did not show protective effects when VEGF was used (117,118). Given the critical role of epicardial progenitor cells in inducing myocardial growth, epicardium-derived growth factor may be another potential target. Epicardial follistatin-like 1 (FSTL1) is normally expressed in the epicardium; the lack of FSTL1 induces inadequate response to injury, whereas FSTL supplementation effectively inhibits myocardial death and reverses remodeling. Interestingly, only epicardium-derived FSTL1 (as opposed to myocardial FSTL1) could stimulate the division of cardiomyocytes, indicating that the source of the signaling molecule and post-translation modifications may play a critical role (119). In a genetic ablation of the epicardium model, the epicardium has been considered a prominent source of mitogens (including NRG1, retinoic acid, and Bmp ligands) to promote tissue repair and vascularization (68,94). Intriguingly, intramyocardial injection of NRG1 using microparticles was found to effectively promote left ventricular function improvement following MI in a porcine study, with reduced ventricular remodeling (120).
In addition to promoting neovascularization, immune modulation could be another potential target to enhance the efficacy of heart regeneration therapy. Although inflammation participates in heart repair after myocardial injury, clinical trials of immunosuppressive agents have failed to obtain positive results, suggesting that the inhibition of inflammation alone may not be an effective means of improving cardiac repair (121). Meanwhile, macrophages have been considered a proregenerative factor in numerous conditions (57,122); the ablation of resident macrophages prevents cardiomyocytes and endothelial cells from proliferating and causes myocardial fibrosis, indicating that embryonic-derived macrophages are a key mediator of cardiac recovery (123). Conversely, infiltrated monocyte-derived macrophages have been identified as a proinflammatory factor inducing a lack of reparative activities following cardiac injury. Thus, it is essential to investigate the roles of immune cell subpopulations in heart regeneration.
Discussion and summary
A few of the prospects for future research on heart regeneration should be mentioned. The generation of reprogrammed cardiomyocytes from human fibroblasts or other cell types will be the next promising strategy for cardiac repair. Ideally, practical cardiovascular regeneration therapy requires better survival of the transplanted iCM to repair the injured myocardium. Moreover, the newly divided cardiomyocytes must couple with the existing muscle to avoid potential arrhythmias. Meanwhile, methods for the transplantation of cardiomyocytes and electromechanical incorporation of local cardiac tissue are also necessary. Importantly, more attention should be paid to the side effects and significant progression of heart regeneration. For instance, the expression of human miR-199a can promote obvious cardiac repair but also causes severe side effects, such as sudden arrhythmic death, due to repeated delivery and excessive miR-199a expression (102). Therefore, the dosage of this therapy needs to be fine-tuned.
With the progression of biomaterials, there is hope that cardiac arrhythmias can be reduced. The implantation of internal cardioverter-defibrillator devices can prevent sudden arrhythmic death. Novel imaging strategies for heart regeneration have prospects for the deep exploration of the complex biological effects as evidence of cell cycle re-entry (124). Heart regeneration is one of the most attractive fields in cardiovascular research and provides a promising future for mending a broken heart and reducing the morbidity and mortality of severe cardiovascular diseases. Although some controversies and challenges remain, a clearer picture of adult mammalian cardiac regeneration is emerging.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82070252 to HM) and the Zhejiang Provincial Natural Science Foundation (No. LR21H020001 to HM).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2649/rc
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2649/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2649/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wencker D, Chandra M, Nguyen K, et al. A mechanistic role for cardiac myocyte apoptosis in heart failure. J Clin Invest 2003;111:1497-504. [Crossref] [PubMed]
- Timmis A, Townsend N, Gale CP, et al. European Society of Cardiology: Cardiovascular Disease Statistics 2019. Eur Heart J 2020;41:12-85. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356-87. [Crossref] [PubMed]
- Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98-102. [Crossref] [PubMed]
- Mollova M, Bersell K, Walsh S, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A 2013;110:1446-51. [Crossref] [PubMed]
- Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev 2007;87:521-44. [Crossref] [PubMed]
- Soonpaa MH, Kim KK, Pajak L, et al. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol 1996;271:H2183-9. [PubMed]
- Walsh S, Pontén A, Fleischmann BK, et al. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res 2010;86:365-73. [Crossref] [PubMed]
- Bergmann O, Jovinge S. Cardiac regeneration in vivo: mending the heart from within? Stem Cell Res 2014;13:523-31. [Crossref] [PubMed]
- Malliaras K, Zhang Y, Seinfeld J, et al. Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol Med 2013;5:191-209. [Crossref] [PubMed]
- Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013;493:433-6. [Crossref] [PubMed]
- Eschenhagen T, Bolli R, Braun T, et al. Cardiomyocyte Regeneration: A Consensus Statement. Circulation 2017;136:680-6. [Crossref] [PubMed]
- Hsieh PC, Segers VF, Davis ME, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 2007;13:970-4. [Crossref] [PubMed]
- Bergmann O, Zdunek S, Felker A, et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015;161:1566-75. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078-80. [Crossref] [PubMed]
- Haubner BJ, Schneider J, Schweigmann U, et al. Functional Recovery of a Human Neonatal Heart After Severe Myocardial Infarction. Circ Res 2016;118:216-21. [Crossref] [PubMed]
- Laflamme MA, Murry CE. Heart regeneration. Nature 2011;473:326-35. [Crossref] [PubMed]
- Yahalom-Ronen Y, Rajchman D, Sarig R, et al. Reduced matrix rigidity promotes neonatal cardiomyocyte dedifferentiation, proliferation and clonal expansion. Elife 2015;4:e07455. [Crossref] [PubMed]
- Santini MP, Forte E, Harvey RP, et al. Developmental origin and lineage plasticity of endogenous cardiac stem cells. Development 2016;143:1242-58. [Crossref] [PubMed]
- Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003;114:763-76. [Crossref] [PubMed]
- Ellison GM, Vicinanza C, Smith AJ, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 2013;154:827-42. [Crossref] [PubMed]
- van Berlo JH, Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med 2014;20:1386-93. [Crossref] [PubMed]
- Li Y, He L, Huang X, et al. Genetic Lineage Tracing of Nonmyocyte Population by Dual Recombinases. Circulation 2018;138:793-805. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A 2013;110:187-92. [Crossref] [PubMed]
- Paige SL, Plonowska K, Xu A, et al. Molecular regulation of cardiomyocyte differentiation. Circ Res 2015;116:341-53. [Crossref] [PubMed]
- González-Rosa JM, Sharpe M, Field D, et al. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell 2018;44:433-446.e7. [Crossref] [PubMed]
- Bersell K, Arab S, Haring B, et al. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 2009;138:257-70. [Crossref] [PubMed]
- Kimura W, Xiao F, Canseco DC, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature 2015;523:226-30. [Crossref] [PubMed]
- Patterson M, Barske L, Van Handel B, et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet 2017;49:1346-53. [Crossref] [PubMed]
- Zebrowski DC, Vergarajauregui S, Wu CC, et al. Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. Elife 2015;4:e05563. [Crossref] [PubMed]
- Ahuja P, Perriard E, Perriard JC, et al. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci 2004;117:3295-306. [Crossref] [PubMed]
- Engel FB, Schebesta M, Keating MT. Anillin localization defect in cardiomyocyte binucleation. J Mol Cell Cardiol 2006;41:601-12. [Crossref] [PubMed]
- Brodsky VYa, Sarkisov DS, Arefyeva AM, et al. Polyploidy in cardiac myocytes of normal and hypertrophic human hearts; range of values. Virchows Arch 1994;424:429-35. [Crossref] [PubMed]
- Quaife-Ryan GA, Sim CB, Ziemann M, et al. Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation 2017;136:1123-39. [Crossref] [PubMed]
- Puente BN, Kimura W, Muralidhar SA, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 2014;157:565-79. [Crossref] [PubMed]
- Kolwicz SC Jr, Purohit S, Tian R. Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 2013;113:603-16. [Crossref] [PubMed]
- Cao T, Liccardo D, LaCanna R, et al. Fatty Acid Oxidation Promotes Cardiomyocyte Proliferation Rate but Does Not Change Cardiomyocyte Number in Infant Mice. Front Cell Dev Biol 2019;7:42. [Crossref] [PubMed]
- Cardoso AC, Lam NT, Savla JJ, et al. Mitochondrial Substrate Utilization Regulates Cardiomyocyte Cell Cycle Progression. Nat Metab 2020;2:167-78. [Crossref] [PubMed]
- Tzahor E, Poss KD. Cardiac regeneration strategies: Staying young at heart. Science 2017;356:1035-9. [Crossref] [PubMed]
- Gu Z, Steinmetz LM, Gu X, et al. Role of duplicate genes in genetic robustness against null mutations. Nature 2003;421:63-6. [Crossref] [PubMed]
- Quaife-Ryan GA, Sim CB, Porrello ER, et al. Resetting the epigenome for heart regeneration. Semin Cell Dev Biol 2016;58:2-13. [Crossref] [PubMed]
- Keating ST, El-Osta A. Epigenetics and metabolism. Circ Res 2015;116:715-36. [Crossref] [PubMed]
- Gilsbach R, Preissl S, Grüning BA, et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun 2014;5:5288. [Crossref] [PubMed]
- Sim CB, Ziemann M, Kaspi A, et al. Dynamic changes in the cardiac methylome during postnatal development. FASEB J 2015;29:1329-43. [Crossref] [PubMed]
- Ounzain S, Micheletti R, Beckmann T, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353-68a. [Crossref] [PubMed]
- Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206-14. [Crossref] [PubMed]
- Klattenhoff CA, Scheuermann JC, Surface LE, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570-83. [Crossref] [PubMed]
- Slepak TI, Webster KA, Zang J, et al. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J Biol Chem 2001;276:7575-85. [Crossref] [PubMed]
- Sartorelli V, Huang J, Hamamori Y, et al. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol 1997;17:1010-26. [Crossref] [PubMed]
- Takaya T, Kawamura T, Morimoto T, et al. Identification of p300-targeted acetylated residues in GATA4 during hypertrophic responses in cardiac myocytes. J Biol Chem 2008;283:9828-35. [Crossref] [PubMed]
- Shikama N, Lutz W, Kretzschmar R, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J 2003;22:5175-85. [Crossref] [PubMed]
- Montgomery RL, Davis CA, Potthoff MJ, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 2007;21:1790-802. [Crossref] [PubMed]
- Montgomery RL, Potthoff MJ, Haberland M, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest 2008;118:3588-97. [Crossref] [PubMed]
- Trivedi CM, Lu MM, Wang Q, et al. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem 2008;283:26484-9. [Crossref] [PubMed]
- Godwin JW, Brockes JP. Regeneration, tissue injury and the immune response. J Anat 2006;209:423-32. [Crossref] [PubMed]
- Sattler S, Rosenthal N. The neonate versus adult mammalian immune system in cardiac repair and regeneration. Biochim Biophys Acta 2016;1863:1813-21. [Crossref] [PubMed]
- Aurora AB, Porrello ER, Tan W, et al. Macrophages are required for neonatal heart regeneration. J Clin Invest 2014;124:1382-92. [Crossref] [PubMed]
- Li J, Tan J, Martino MM, et al. Regulatory T-Cells: Potential Regulator of Tissue Repair and Regeneration. Front Immunol 2018;9:585. [Crossref] [PubMed]
- Witman N, Murtuza B, Davis B, et al. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev Biol 2011;354:67-76. [Crossref] [PubMed]
- Laube F, Heister M, Scholz C, et al. Re-programming of newt cardiomyocytes is induced by tissue regeneration. J Cell Sci 2006;119:4719-29. [Crossref] [PubMed]
- González-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat Protoc 2012;7:782-8. [Crossref] [PubMed]
- Wang J, Panáková D, Kikuchi K, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011;138:3421-30. [Crossref] [PubMed]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188-90. [Crossref] [PubMed]
- Darehzereshki A, Rubin N, Gamba L, et al. Differential regenerative capacity of neonatal mouse hearts after cryoinjury. Dev Biol 2015;399:91-9. [Crossref] [PubMed]
- Ye L, D'Agostino G, Loo SJ, et al. Early Regenerative Capacity in the Porcine Heart. Circulation 2018;138:2798-808. [Crossref] [PubMed]
- Haubner BJ, Adamowicz-Brice M, Khadayate S, et al. Complete cardiac regeneration in a mouse model of myocardial infarction. Aging (Albany NY) 2012;4:966-77. [Crossref] [PubMed]
- Chablais F, Veit J, Rainer G, et al. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol 2011;11:21. [Crossref] [PubMed]
- Wang J, Cao J, Dickson AL, et al. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015;522:226-30. [Crossref] [PubMed]
- Kikuchi K, Holdway JE, Werdich AA, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601-5. [Crossref] [PubMed]
- Jopling C, Sleep E, Raya M, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606-9. [Crossref] [PubMed]
- van Berlo JH, Kanisicak O, Maillet M, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 2014;509:337-41. [Crossref] [PubMed]
- Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 2015;33:155-60. [Crossref] [PubMed]
- DeLaughter DM, Bick AG, Wakimoto H, et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 2016;39:480-90. [Crossref] [PubMed]
- Skelly DA, Squiers GT, McLellan MA, et al. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep 2018;22:600-10. [Crossref] [PubMed]
- Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015;161:1202-14. [Crossref] [PubMed]
- Kretzschmar K, Watt FM. Lineage tracing. Cell 2012;148:33-45. [Crossref] [PubMed]
- Zong H, Espinosa JS, Su HH, et al. Mosaic analysis with double markers in mice. Cell 2005;121:479-92. [Crossref] [PubMed]
- Nguyen NUN, Canseco DC, Xiao F, et al. A calcineurin-Hoxb13 axis regulates growth mode of mammalian cardiomyocytes. Nature 2020;582:271-6. [Crossref] [PubMed]
- Magadum A, Singh N, Kurian AA, et al. Pkm2 Regulates Cardiomyocyte Cell Cycle and Promotes Cardiac Regeneration. Circulation 2020;141:1249-65. [Crossref] [PubMed]
- El-Nachef D, Bugg D, Beussman KM, et al. Engrafted Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Undergo Clonal Expansion In Vivo. Circulation 2021;143:1635-8. [Crossref] [PubMed]
- Sereti KI, Nguyen NB, Kamran P, et al. Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat Commun 2018;9:754. [Crossref] [PubMed]
- Velagaleti RS, Pencina MJ, Murabito JM, et al. Long-term trends in the incidence of heart failure after myocardial infarction. Circulation 2008;118:2057-62. [Crossref] [PubMed]
- He L, Nguyen NB, Ardehali R, et al. Heart Regeneration by Endogenous Stem Cells and Cardiomyocyte Proliferation: Controversy, Fallacy, and Progress. Circulation 2020;142:275-91. [Crossref] [PubMed]
- Jensen B, van den Berg G, van den Doel R, et al. Development of the hearts of lizards and snakes and perspectives to cardiac evolution. PLoS One 2013;8:e63651. [Crossref] [PubMed]
- Jacyniak K, Vickaryous MK. Constitutive cardiomyocyte proliferation in the leopard gecko (Eublepharis macularius). J Morphol 2018;279:1355-67. [Crossref] [PubMed]
- Xiao J, Liu H, Cretoiu D, et al. miR-31a-5p promotes postnatal cardiomyocyte proliferation by targeting RhoBTB1. Exp Mol Med 2017;49:e386. [Crossref] [PubMed]
- Sudi S, Chin YZ, Wasli NS, et al. Carpaine Promotes Proliferation and Repair of H9c2 Cardiomyocytes after Oxidative Insults. Pharmaceuticals (Basel) 2022;15:230. [Crossref] [PubMed]
- Bicknell KA, Coxon CH, Brooks G. Forced expression of the cyclin B1-CDC2 complex induces proliferation in adult rat cardiomyocytes. Biochem J 2004;382:411-6. [Crossref] [PubMed]
- Chaudhry HW, Dashoush NH, Tang H, et al. Cyclin A2 mediates cardiomyocyte mitosis in the postmitotic myocardium. J Biol Chem 2004;279:35858-66. [Crossref] [PubMed]
- Pasumarthi KB, Nakajima H, Nakajima HO, et al. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res 2005;96:110-8. [Crossref] [PubMed]
- Di Stefano V, Giacca M, Capogrossi MC, et al. Knockdown of cyclin-dependent kinase inhibitors induces cardiomyocyte re-entry in the cell cycle. J Biol Chem 2011;286:8644-54. [Crossref] [PubMed]
- Mahmoud AI, Kocabas F, Muralidhar SA, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 2013;497:249-53. [Crossref] [PubMed]
- Schindler YL, Garske KM, Wang J, et al. Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 2014;141:3112-22. [Crossref] [PubMed]
- Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development 2016;143:729-40. [Crossref] [PubMed]
- Uygur A, Lee RT. Mechanisms of Cardiac Regeneration. Dev Cell 2016;36:362-74. [Crossref] [PubMed]
- Engel FB, Hsieh PC, Lee RT, et al. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A 2006;103:15546-51. [Crossref] [PubMed]
- Heallen T, Morikawa Y, Leach J, et al. Hippo signaling impedes adult heart regeneration. Development 2013;140:4683-90. [Crossref] [PubMed]
- Bassat E, Mutlak YE, Genzelinakh A, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 2017;547:179-84. [Crossref] [PubMed]
- Morikawa Y, Heallen T, Leach J, et al. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature 2017;547:227-31. [Crossref] [PubMed]
- Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376-81. [Crossref] [PubMed]
- Tao G, Kahr PC, Morikawa Y, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature 2016;534:119-23. [Crossref] [PubMed]
- Gabisonia K, Prosdocimo G, Aquaro GD, et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 2019;569:418-22. [Crossref] [PubMed]
- Ogle BM, Bursac N, Domian I, et al. Distilling complexity to advance cardiac tissue engineering. Sci Transl Med 2016;8:342ps13. [Crossref] [PubMed]
- Chong JJ, Yang X, Don CW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014;510:273-7. [Crossref] [PubMed]
- Shiba Y, Gomibuchi T, Seto T, et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016;538:388-91. [Crossref] [PubMed]
- Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010;142:375-86. [Crossref] [PubMed]
- Srivastava D, DeWitt N. In Vivo Cellular Reprogramming: The Next Generation. Cell 2016;166:1386-96. [Crossref] [PubMed]
- Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012;485:593-8. [Crossref] [PubMed]
- Fu JD, Srivastava D. Direct reprogramming of fibroblasts into cardiomyocytes for cardiac regenerative medicine. Circ J 2015;79:245-54. [Crossref] [PubMed]
- Hashimoto H, Wang Z, Garry GA, et al. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell 2019;25:69-86.e5. [Crossref] [PubMed]
- Lalit PA, Salick MR, Nelson DO, et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 2016;18:354-67. [Crossref] [PubMed]
- Cao N, Huang Y, Zheng J, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016;352:1216-20. [Crossref] [PubMed]
- Jayawardena T, Mirotsou M, Dzau VJ. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol Biol 2014;1150:263-72. [Crossref] [PubMed]
- Zhou Y, Wang L, Vaseghi HR, et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016;18:382-95. [Crossref] [PubMed]
- Christoforou N, Chellappan M, Adler AF, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One 2013;8:e63577. [Crossref] [PubMed]
- Zangi L, Lui KO, von Gise A, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 2013;31:898-907. [Crossref] [PubMed]
- Giacca M, Zacchigna S. VEGF gene therapy: therapeutic angiogenesis in the clinic and beyond. Gene Ther 2012;19:622-9. [Crossref] [PubMed]
- Taimeh Z, Loughran J, Birks EJ, et al. Vascular endothelial growth factor in heart failure. Nat Rev Cardiol 2013;10:519-30. [Crossref] [PubMed]
- Wei K, Serpooshan V, Hurtado C, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature 2015;525:479-85. [Crossref] [PubMed]
- Garbayo E, Gavira JJ, de Yebenes MG, et al. Catheter-based Intramyocardial Injection of FGF1 or NRG1-loaded MPs Improves Cardiac Function in a Preclinical Model of Ischemia-Reperfusion. Sci Rep 2016;6:25932. [Crossref] [PubMed]
- Ruparelia N, Chai JT, Fisher EA, et al. Inflammatory processes in cardiovascular disease: a route to targeted therapies. Nat Rev Cardiol 2017;14:133-44. [Crossref] [PubMed]
- Eming SA, Wynn TA, Martin P. Inflammation and metabolism in tissue repair and regeneration. Science 2017;356:1026-30. [Crossref] [PubMed]
- Lavine KJ, Epelman S, Uchida K, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A 2014;111:16029-34. [Crossref] [PubMed]
- Naumova AV, Modo M, Moore A, et al. Clinical imaging in regenerative medicine. Nat Biotechnol 2014;32:804-18. [Crossref] [PubMed]
(English Language Editors: A. Kassem and J. Gray)