
Systematic analysis of microbiota in pregnant Chinese women and its association with miscarriage
Introduction
Miscarriage is the most common adverse pregnancy outcome and affects approximately 15% of clinically diagnosed pregnancies (1-4). After ruling out factors such as parental chromosomal anomalies, maternal thrombophilic disorders, immune dysfunction, and various endocrine disturbances, more than 50% of miscarriage cases remain unexplained (5,6). There have been many possible causes of miscarriage reported in earlier studies using non-objective, symptomatic measures. A rare cause of pregnancy loss is infection (7); however, the mechanism(s) of infection-induced miscarriage are not yet fully elucidated (8,9).
The maternal microbiota (occupying various body sites such as the gut, vagina, and cervix) both influences and is affected by physiological and pathological changes in pregnancy (10). In fact, previous research suggests an interactive relationship between microbiome perturbation and immune dysregulation. Thus, the immune maybe activated when the microbiota and its metabolites was changed. In turn, chronic inflammation may shape the structure and function of the microbial communities. The normal reproductive tract flora predominantly consists of Lactobacillus species bacteria in healthy women (11), and low bacterial richness and diversity (12,13). Some researchers have found that first trimester miscarriage was associated with reduced prevalence of Lactobacillus spp. dominated vaginal microbiota and increased bacterial diversity and richness (13), yet others have found that the decreased bacterial diversity as well as alteration of the composition of gut microbiota were associated with early miscarriage (14). These discrepancies could be attributed to the study populations of different racial and ethnic groups, variety of the sampling methods and sampling sites, and variation in the time of sampling during the pregnancy (at different gestational ages) (15,16). Due to the complex etiology and pathogenesis of miscarriage, the association of microbiota and miscarriage remains insufficiently understood and more studies are needed (17).
Given that evidence has demonstrated the microbe-host interactions during pregnancy (18), we hypothesized that the association of host microbiota and miscarriage may be modified by host heterogeneity [e.g., gestational age, body mass index (BMI), parity, and previous miscarriage]. As different sites within and upon the human body (e.g., the skin, mouth, nasal cavity, gut, and reproductive tract) harbor discrete populations of microbes, we also hypothesized that the host microbial communities from different body parts may interact and present different pathways for miscarriage. Rectal swabs appeared to be a convenient means of sampling the human gut microbiota (19), so we collected samples from the vagina, cervix, and the rectum, serving as a proxy to the profiling of gut microbiota. In the current study, we aimed to use 16S ribosomal RNA (rRNA) amplification and metagenomic sequencing technology to characterize the bacterial composition, taking into account the host heterogeneity by performing multi-omics analyses, of Chinese women that had experienced first trimester miscarriages versus controls who voluntarily underwent elective abortion. An understanding of the association between miscarriage and the microecology during pregnancy could lead to precision intervention for women of childbearing age, and reduce the occurrence of undesirable pregnancy outcomes (20). We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4115/rc).
Methods
Study design and sample collection
This study was a case-control study conducted at the Shenzhen Dapeng New District Maternal and Child Health Hospital between January 2019 and May 2020. The women in the first trimester of pregnancy with a missed miscarriage who visited for surgery were enrolled in this study. Women who elected to undergo an abortion during the same period were included as controls and were matched to cases for maternal age, fertility story (gravity and parity), and BMI. A missed miscarriage pregnancy was determined by a uterus size that remained smaller than that of the gestational weeks, elevated urine or blood human chorionic gonadotropin (HCG), menstruation, gestational sac confirmed by ultrasound but smaller than that of the gestational weeks, no embryo visible in the sac, or an undetectable heartbeat. Participants with comorbidities of acute and chronic infectious diseases, preeclampsia, gestational diabetes, and antibiotic, probiotic, or glucocorticoid use within 6 months were excluded.
The vaginal, cervical, and rectal samples were collected using standard the operation procedures developed by the Shenzhen Dapeng New District Maternal and Child Health Hospital: (I) before surgery—disinfect the vulva, and expose the orificium externum uteri to the speculum; and (II) during surgery—the vaginal secretions were scraped from the vaginal walls of each participant using sterile, cotton-tipped swabs. To avoid contamination, the swabs did not touch the vaginal wall or other structures (vagina). Immediately after, extending 1 cm into the cervical canal, by a second sterile cotton-tipped swab was applied to obtain a cervical secretion sample (cervix). After sterilizing the anal orifice, a third sterile cotton-tipped swab was extended 3 cm into the rectum to obtain the rectal samples (rectum). After sampling, swabs were placed in separate sterile cryopreservation tubes immediately and stored at −80 ℃ within 20 minutes of collection. Samples were then transported on dry ice to the Laboratory of CheerLand Institute of Precision Medicine in Shenzhen for further sequencing and analysis.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Shenzhen Dapeng New District Maternal and Child Health Hospital (No. FY201801), and informed consent was provided by all participants before enrollment in the project.
Clinical and demographic data
Blood and urine samples were routinely obtained from the pregnant women on the same day as the specimen sample collection (surgery). The demographic information of all the participants was collected through self-reporting at the first clinic visit (9–12 weeks of gestation) or during their first visit to the hospital (whichever was earlier). Participants’ BMI, gestational disease status, pregnancy outcomes, and neonatal information were obtained from their hospital electronic medical records.
Extracting samples DNA and 16S rRNA gene amplicon sequencing
We used next generation sequencing technology to sequence the 16s rDNA amplicon of microbes from the three sample sites (21). These total DNA of microbe of samples was extracted by QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany). When these total DNA samples passed the quality control of nanodrop (Thermo Fisher Scientific, Waltham, MA, USA), Qubit 3.0 (Invitrogen, Carlsbad, CA, USA), and agarose gel electrophoresis, we used these total DNA samples as a template to amplify the 16S rRNA V4 region. Using the amplicon polymerase chain reaction (PCR) technology, the 16S rRNA gene V4 region of total DNA was amplified by universal primers V4-515F (5'-GTGCCAGCMGCCGCGGTAA-3') and 806R (5'- GGACTACHVGGGTWTCTAAT-3'). We detected and purified the amplicon product using agarose gel electrophoresis. The products of which the concentration was qualified underwent a series of operations such as end repair and adapter ligation to acquire sequencing library. Quality control was conducted using a Qubit 3.0 and an Agilent 2100 analyzer (Agilent, Santa, Clara, CA, USA), and the sequencing library was sequenced by the Illumina Mini Seq platform (Illumina, San Diego, CA, USA) for PE150 sequencing.
Bioinformatic analysis
We used the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to process the raw data to product clean sequencing data. Using a QIIME2 standard analysis pipeline, the clean sequencing data was treated, including trimming of the sequencing primer and barcodes, removing chimeric artifacts, clustering operational taxonomic units (OTUs), picking OTU reference sequences, and annotating taxonomic information (22,23). In the clustering OTUs step, the clean sequencing data were clustered into OTUs based on 97% similarity using the UCLUST program of QIIME2. The OTU reference sequences were aligned against the Silva 138 SSURef NR99 database (24) to annotate taxonomic information using the UCLUST taxonomy assigner. We quantified phylum and genus relative abundance by as a ratio of the phylum (genus) OTUs reads number to the total reads number.
Statistical analysis
Differences in bacterial richness (species observed) and diversity (Shannon index) between groups were analyzed using t-test or Mann-Whitney test where appropriate, for each sample site. Differences in bacterial richness and diversity among samples from different sites were tested using paired t-test or Wilcoxon signed rank test where appropriate. Multivariable regression analyses on the bacterial diversity were performed taking into account the host factors and clinical measures to further explore the difference in microbiota between two groups using Stata software (v14.0; StataCorp., College Station, TX, USA). All significance levels were set at P<0.05.
Results
Description of the study participants
The basic characteristics of all participants are summarized in Table 1. No statistical difference was observed in maternal age, number of pregnancies, number of live births, and BMI between the case and control groups (all P>0.05). However, women in the control group had no previous history missed miscarriage yet over 85% of those with missed miscarriage had experienced one missed miscarriage before. Women who had undergone a voluntary elective abortion underwent surgery earlier in the first trimester than those with missed miscarriage. Table 2 shows the clinical measures of all participants at the time of the surgery. There was an apparent trend that more women in the case group had abnormal clinical measures than controls, but the difference was not statistically significant. Due to the uneven number between two groups, the potential interferences of the host factors could not be eliminated.
Table 1
| Characteristics | Missed miscarriage (n=63) | Elective abortion (n=24) | P value |
|---|---|---|---|
| Maternal age years, mean (SD)* | 30.6 (6.0) | 30.0 (6.1) | 0.69 |
| Pre-pregnancy weight (kg), mean (SD)* | 54.0 (8.1) | 50.0 (3.6) | 0.01 |
| BMI (kg/m2), mean (SD)* | 21.1 (3.1) | 20.3 (1.6) | 0.25 |
| Fertility history, n (%) | |||
| Gravity^ | 0.57 | ||
| Primigravid | 15 (23.8) | 4 (16.7) | |
| Multigravid | 48 (76.2) | 20 (83.3) | |
| Parity^ | 0.18 | ||
| Nulliparous | 21 (33.3) | 4 (16.7) | |
| Multiparous | 42 (66.7) | 20 (83.3) | |
| Previous missed miscarriage, n (%)^ | <0.001 | ||
| 0 | 0 | 24 (100.0) | |
| 1 | 54 (85.7) | 0 | |
| ≥2 | 9 (14.3) | 0 | |
| Pregnancy complication, yes (%)# | 23 (36.5) | 7 (29.2) | 0.52 |
| Gestational week (continuous) | 8.7 (1.9) | 7.2 (2.3) | 0.002 |
| Gestational week# | 0.01 | ||
| <8 | 28 (44.4) | 18 (75.0) | |
| 8–12 | 35 (55.6) | 6 (25.0) |
*, significance was tested by t-test; ^, significance was tested by Fisher’s exact; #, significance was tested by chi-square test. BMI, body mass index.
Table 2
| Abnormal clinical measures | Missed miscarriage (N=63), n (%) | Elective abortion (N=24), n (%) | P value |
|---|---|---|---|
| RBC | 3 (4.8) (n=62) | 2 (8.7) (n=23) | 0.61 |
| WBC | 3 (4.8) (n=62) | 4 (17.4) (n=23) | 0.08 |
| ALT | 3 (5.2) (n=58) | 0 (0) (n=9) | 0.99 |
| UA | 4 (6.9) (n=58) | 1 (11.1) (n=9) | 0.53 |
| Urea | 0 (0) (n=58) | 0 (0) (n=9) | |
| AST | 1 (2.0) (n=51) | 0 (0) (n=9) | 0.99 |
| GLU | 6 (11.8) (n=51) | 0 (0) (n=9) | 0.58 |
| SBP | 15 (26.3) (n=57) | 2 (28.6) (n=7) | 0.99 |
| DBP | 9 (15.8) (n=57) | 0 (0) (n=7) | 0.58 |
| TBIL | 7 (14.0) (n=50) | 0 (0) (n=5) | 0.99 |
RBC, red blood count; WBC, white blood count; ALT, alanine transaminase; UA, uric acid; AST, aspartate transaminase; GLU, glucose; SBP, systolic blood pressure; DBP, diastolic blood pressure; TBIL, total bilirubin.
Decreased bacterial diversity in rectum and cervix, but not in vagina was associated with miscarriage patients
A total of 261 swab samples were sequenced, providing an average of 761,638 reads per sample per sample site, and the sequencing data of these samples were clustered to 289 OTUs. According to sample site, compared to the rectal samples, the reproductive tract samples (vagina and cervix) displayed significantly lower alpha-diversity in general (Figure 1A). The Shannon indexes for samples from the rectum and cervix were significantly decreased in the missed miscarriage (case) group relative to the elective abortion (control) group (Figure 1B), indicating a lower richness and evenness of rectal and cervical bacteria in women with missed miscarriage. However, there was no statistically significant difference in vaginal bacterial diversity between the two groups. We evaluated the differences in microbiome community for three sites between the two groups using beta diversity and visualized analysis via principal coordinate analysis (PCoA) plots. We could readily distinguish the reproductive tract samples (vagina and cervix) and rectum samples by PCoA based on their general profiles (Figure 1C). Additionally, the weighted PCoA plots revealed that the microbiota in participants who had miscarried was not significantly clustered compared to that of the control participants (Figure 1D-1F).

Alterations in the composition of rectal and reproductive tract microbiota associated with miscarriage
The relative abundance of taxa in the different groups was utilized to investigate the significant changes of the microbiota in the samples. Comprehensively, the rectal microbiota of pregnant women contains five major bacterial phyla: Firmicutes (accounting for 79.2% and 72.5% of OTUs in the missed miscarriage and control groups, respectively) and Bacteroidetes (9% and 14.6%) (Figure 2A), followed by Actinobacteria (8.6% and 7.1%), Proteobacteria (2.2% and 4.2%), and Fusobacteria (0.4% and 0.9%). This compositional pattern of bacterial phyla was slightly different from that seen in the cervix and vagina microbiota, which were dominated by Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria, Tenericutes, and Proteobacteria (Figure 2A). Firmicutes of the rectal OTUs was enriched in the missed miscarriage group compared to the control group (P<0.001), whereas Proteobacteria was enriched in the control group (4.2% vs. 2.2% in missed miscarriage group, P=0.001). The Firmicutes/Bacteroidetes ratio has been suggested as an indicator of several pathological conditions (25). The ratio was consistently higher in the missed miscarriage group than that in the control group, across rectal OTUs (2.5 and 1.7 on Log10, P=0.003) and cervical OTUs (4.7 and 3.9, P=0.06), but not in vaginal OTUs (4.5 and 4.3, P=0.67), indicating that a pathological change occurred in women with missed miscarriage (Figure 2B).
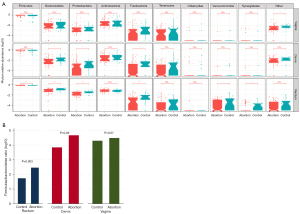
Figure 3A-3C shows a heatmap of the species abundance in two groups in different sites of every individual. In Figure 3A,3B, regarding the vaginal and cervical microbiome, the most abundant genus found in both groups was Lactobacillus. We identified four major bacteria in the vagina, including Lactobacillus, Gardnerella, Atopobium, and Prevotella. Of all the detected genera, Lactobacillus of the phylum Firmicutes were the most abundant. The relative abundance of Lactobacillus was low. The enrichment of facultative or strict anaerobic species, such as Gardnerella or Atopobium, was often associated with bacterial vaginosis (BV). The main genera found in the rectum were Anaerococcus, Peptoniphilus, Lactobacillus, Finegoldia, and Faecalibacterium (Figure 3C).
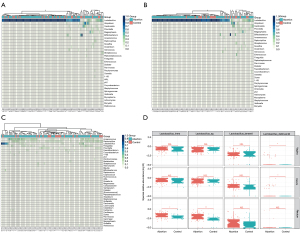
We observed that the Lactobacillus from vaginal and cervix samples was significantly more abundant in the case group than that in control group (P<0.002 and P<0.013, respectively). However, the difference was not significant after adjustment for host factors including age, BMI, gestational age, previous missed miscarriage, and normal red blood count (RBC) and white blood count (WBC; Table S1). This indicated that missed miscarriage may be independent of the presence of Lactobacillus-dominated microbiota communities, and the association was modified by the host factors and clinical measures. Figure 3D shows the relative abundances of the top 4 most abundant Lactobacillus in the three sampling sites. In the vaginal samples, Lactobacillus delbrueckii was significantly more abundant in the case group than in the control group (Figure 3D).
Association of host factors and microbiota
Multivariable analysis revealed that host factors did not consistently impact the diversity of the microbiota, and the effect size was varied. The significant effect was only seen in the association of rectal microbiota in our model (Table 3). Gestational age at the time of surgery was positively associated with the diversity of rectal microbiota, with an effect size of 10% (P=0.004). The RBC was negatively associated with the rectum microbiota diversity but with a small effect size of 7.2% (P=0.02). Further analysis of the most predominant phylum from rectal OTUs showed that the difference between groups was mostly independent from host factors, except for Firmicutes, where maternal age had a small effect of 7.8% (P=0.01).
Table 3
| Host factors | Rectum (%) | Cervix (%) | Vagina (%) | Dominated phylum in rectum samples (%) | ||
|---|---|---|---|---|---|---|
| Firmicutes | Bacteriodetes | Actinobacteria | ||||
| Group^ | 7.3* | 5.2* | 1.00 | 18.1*** | 18.7*** | 12.3** |
| Model# | 22.5** | 6.77 | 3.04 | 27.4*** | 23.1** | 17.8* |
| Group | 8.9* | 1.87 | 0.29 | 11.6** | 10.2** | 6.0* |
| Age | 0.00 | 0.17 | 0.02 | 7.8* | 2.10 | 1.50 |
| BMI | 0.80 | 0.34 | 0.90 | 1.30 | 0.04 | 1.80 |
| Gestational week | 10.0** | 0.06 | 0.00 | 0.10 | 0.20 | 1.60 |
| Previous miscarriage | 2.00 | 0.11 | 0.01 | 1.60 | 1.00 | 0.20 |
| WBC | 0.20 | 1.15 | 0.61 | 0.30 | 0.09 | 0.80 |
| RBC | 7.2* | 0.75 | 0.62 | 0.90 | 0.50 | 0.30 |
^, include only one variable ‘group’ (case and control) in the model, the effect size of the variable equals to the effect size of the entire model; #, include all the variables into the model. Effect size was estimated from the coefficient. *, P<0.05; **, P <0.01; ***, P<0.001. BMI, body mass index; RBC, red blood count; WBC, white blood count.
Discussion
We evaluate the microbial community along the female reproductive tract and rectum of women with missed miscarriage compared with those who underwent voluntary elective abortion by 16S rRNA sequencing. In this study, we found significant changes in the diversity and composition of rectal microbiota in women with abortion. The reduce of gut microbial diversity may means a higher risk of gastrointestinal disease and inflammation (26,27). A previous study (28) also reported that chronic inflammation was related to an increased ratio of Firmicutes to Bacteroidetes. Cytokines are produced by the inflammatory response, and the cytokines proteins protect the body against infection and interfere with iron processing and red blood cell production (29). Severe inflammation can cause internal bleeding that leads to a decrease in RBC (30). Here we reported a negative association between RBC and rectal microbiota diversity, indicating a relationship between rectal microbiota and the increased inflammatory activities in women who experience missed miscarriage. In our study, we observed both reduced rectal microbiota and an increased ratio of Firmicutes to Bacteriodetes in the miscarriage group, consistent with a previous observation (14), indicating that the inflammatory effects of the rectal microbiome in women with missed miscarriage are more likely to be caused by holistic dysbiosis than a specific pathogen.
The Lactobacillus can protect the vagina, L. Crispatus is important for maintaining the stability of the vaginal environment in pregnant women on the other hand, high estrogen can induce lactobacilli to decompose glycogen and lactic acid.by the way, the low vaginal pH also beneficial to the production of Lactobacillus and protection from harmful bacteria. so, vaginal pH can be a predictive index of vaginal infection (31). vaginal samples deplete of Lactobacillus displayed increased richness and diversity and colonization by potential pathogens including Prevotella, Megasphaera, Anaerococcus, and Peptoniphilus genus. Other recent studies have also identified an association between vaginal Lactobacillus spp. depletion and adverse reproductive health outcomes (32,33). In our findings, the vaginal and cervical samples were also dominated by Lactobacillus spp., followed by Atopobium and Gardnerella. A study has reported that many factors could affect the stability of the vaginal microflora, including that different species of Lactobacillus could be change the vaginal microbial community. Lactobacillus crispatus could be maintain the stability of the vaginal environment throughout pregnancy. However, if Lactobacillus gasseri and/or Lactobacillus iners dominate during the first trimester, they can induce abnormal vaginal bacterial conditions from the third trimester onwards (34). However, Lactobacillus crispatus was not found in our samples, and the most abundant species was Lactobacillus iners in our study. Yang et al. (18) have extensively explored the association between the gut microbiota of pregnant women and its relationship with host factors. They found that individual heterogeneity is the major factor which shapes the gut microbiome during pregnancy. Our findings were consistent with those of Yang et al.: the gut microbiota is associated with several host factors, such as age and gestational weeks.
In conclusion, we have described the characteristics of the microflora composition in the reproductive tract and rectum of women with missed miscarriage and compared it to those of women who underwent elective abortion. We found a significantly lower rectal microbiota diversity and a proinflammatory tendency in the miscarriage group. We observed a protective role of Lactobacillus flora in the cervix and vagina against miscarriage. Unlike other causes of miscarriage, the vaginal and cervical microbiomes can be potentially modifiable by treatment, such as the use of targeted antibiotic, prebiotic, or probiotic treatments, bacteriophage, and other novel therapies. Further studies are warranted to further explore the role of microbiota communities in miscarriage and how treatment strategies might help us to prevent its occurrence. This study provides an insight into the first-trimester reproductive system and rectal microbiota composition and its relationship with miscarriage. There is evidence indicate that the microbiota of the maternal gut and vagina not only affects the colonization of the infants, but also plays a role before delivery, thereby affecting the long-term immune development of the infants, this implies that the maternal microbiome plays an important role in infant immunity and disease susceptibility before delivery. In the future, we will conduct more in-depth research to clarify with larger cohort studies whether the certain composition of microbiota does increase miscarriage risk and whether screening of newly pregnant women to improve their flora could prevent adverse pregnancy outcomes.
Acknowledgments
The authors thank the Southern University of Science and Technology-CheerLand Institute of Precision Medicine for providing research equipment.
Funding: This study was funded by grants from a special fund for Dapeng New District Industry Development Special Fund Research Projects (No. YL201800201); Dapeng New District Science and Technology Innovation and Industrial Development Special Fund Project (No. YLKY202101-04); Shenzhen Dapeng New District Medical and Health Group Medical and Health Research Project (Nos. 2020JTYM04, 2021JTYM01); and Shenzhen Second People’s Hospital Clinical Research Project (No. 20203357025).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4115/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4115/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4115/coif). ST, CY, CC, GL, JC, XC and WL report that they were employed by CheerLand Biological Technology Co., Ltd. during the study, and they promise to avoid any conflicts of interest (even superficial conflicts) with the company. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Shenzhen Dapeng New District Maternal and Child Health Hospital (FY201801), and informed consent was provided by all participants before enrollment in the project.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wilcox AJ, Weinberg CR, O'Connor JF, et al. Incidence of early loss of pregnancy. N Engl J Med 1988;319:189-94. [Crossref] [PubMed]
- Wood SL, Brain PH. Medical management of missed abortion: a randomized clinical trial. Obstet Gynecol 2002;99:563-6. [Crossref] [PubMed]
- Wang X, Chen C, Wang L, et al. Conception, early pregnancy loss, and time to clinical pregnancy: a population-based prospective study. Fertil Steril 2003;79:577-84. [Crossref] [PubMed]
- Quenby S, Gallos ID, Dhillon-Smith RK, et al. Miscarriage matters: the epidemiological, physical, psychological, and economic costs of early pregnancy loss. Lancet 2021;397:1658-67. [Crossref] [PubMed]
- Larsen EC, Christiansen OB, Kolte AM, et al. New insights into mechanisms behind miscarriage. BMC Med 2013;11:154. [Crossref] [PubMed]
- Rajcan-Separovic E. Next generation sequencing in recurrent pregnancy loss-approaches and outcomes. Eur J Med Genet 2020;63:103644. [Crossref] [PubMed]
- Simpson JL, Gray RH, Queenan JT, et al. Further evidence that infection is an infrequent cause of first trimester spontaneous abortion. Hum Reprod 1996;11:2058-60. [Crossref] [PubMed]
- Giakoumelou S, Wheelhouse N, Cuschieri K, et al. The role of infection in miscarriage. Hum Reprod Update 2016;22:116-33. [Crossref] [PubMed]
- Campisciano G, Florian F, D'Eustacchio A, et al. Subclinical alteration of the cervical-vaginal microbiome in women with idiopathic infertility. J Cell Physiol 2017;232:1681-8. [Crossref] [PubMed]
- Nuriel-Ohayon M, Neuman H, Koren O. Microbial Changes during Pregnancy, Birth, and Infancy. Front Microbiol 2016;7:1031. [Crossref] [PubMed]
- Suzumori N, Sugiura-Ogasawara M. Genetic factors as a cause of miscarriage. Curr Med Chem 2010;17:3431-7. [Crossref] [PubMed]
- MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep 2015;5:8988. [Crossref] [PubMed]
- Al-Memar M, Bobdiwala S, Fourie H, et al. The association between vaginal bacterial composition and miscarriage: a nested case-control study. BJOG 2020;127:264-74. [Crossref] [PubMed]
- Liu Y, Chen H, Feng L, et al. Interactions between gut microbiota and metabolites modulate cytokine network imbalances in women with unexplained miscarriage. NPJ Biofilms Microbiomes 2021;7:24. [Crossref] [PubMed]
- Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012;150:470-80. [Crossref] [PubMed]
- Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One 2012;7:e36466. [Crossref] [PubMed]
- Zhang F, Zhang T, Ma Y, et al. Alteration of vaginal microbiota in patients with unexplained recurrent miscarriage. Exp Ther Med 2019;17:3307-16. [Crossref] [PubMed]
- Yang H, Guo R, Li S, et al. Systematic analysis of gut microbiota in pregnant women and its correlations with individual heterogeneity. NPJ Biofilms Microbiomes 2020;6:32. [Crossref] [PubMed]
- Budding AE, Grasman ME, Eck A, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One 2014;9:e101344. [Crossref] [PubMed]
- Kroon SJ, Ravel J, Huston WM. Cervicovaginal microbiota, women's health, and reproductive outcomes. Fertil Steril 2018;110:327-36. [Crossref] [PubMed]
- Ye Y. Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2011;2010:153-7. [PubMed]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335-6. [Crossref] [PubMed]
- Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852-7. [Crossref] [PubMed]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6. [Crossref] [PubMed]
- Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027-31. [Crossref] [PubMed]
- Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178-84. [Crossref] [PubMed]
- Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn's disease. Gut 2017;66:813-22. [Crossref] [PubMed]
- Masumoto S, Terao A, Yamamoto Y, et al. Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci Rep 2016;6:31208. [Crossref] [PubMed]
- Weiss G. Iron metabolism in the anemia of chronic disease. Biochim Biophys Acta 2009;1790:682-93. [Crossref] [PubMed]
- Mozos I. Mechanisms linking red blood cell disorders and cardiovascular diseases. Biomed Res Int 2015;2015:682054. [Crossref] [PubMed]
- Mei C, Yang W, Wei X, et al. The Unique Microbiome and Innate Immunity During Pregnancy. Front Immunol 2019;10:2886. [Crossref] [PubMed]
- Kindinger LM, Bennett PR, Lee YS, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome 2017;5:6. [Crossref] [PubMed]
- Eckert LO, Moore DE, Patton DL, et al. Relationship of vaginal bacteria and inflammation with conception and early pregnancy loss following in-vitro fertilization. Infect Dis Obstet Gynecol 2003;11:11-7. [Crossref] [PubMed]
- Verstraelen H, Verhelst R, Claeys G, et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol 2009;9:116. [Crossref] [PubMed]
(English Language Editor: J. Jones)


