
Identification of a novel signature with prognostic value in triple-negative breast cancer through clinico-transcriptomic analysis
Introduction
Breast cancer is the most common malignancy and the second leading cause of death among females (1). Triple-negative breast cancer (TNBC), which accounts for approximately 15% of malignant breast neoplasms, is characterized by the lack of expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) (2-4). When compared with other subtypes, TNBC is perceived as exhibiting a highly aggressive phenotype, with higher grade, larger tumor size, and younger age of onset (4-6). Chemotherapy is currently the mainstay of treatment and improves the prognosis for TNBC to a certain extent (2,7). Nevertheless, the clinical outcomes remain unfavorable with great potential for metastasis and an increased likelihood of death from TNBC (5,7,8).
According to different classifications, including gene expression, metabolic pathways, or proteomic data, each presenting a unique gene expression profiling, metabolic characteristics, or proteomic mapping, TNBC can be further divided into 4, 6, or more subtypes (9-13). The 3-year overall survival (OS) rate is reported to be 68–94% (7). Accumulated evidence has indicated the considerable heterogeneity of the disease, making it challenging to predict prognosis accurately based on clinicopathological factors. Therefore, it is of clinical importance to develop a TNBC-related prognostic tool integrating the genomic signature and conventional clinicopathological parameters for accurate prognosis prediction.
Although an increasing number of breast cancer-related signatures have been established based on transcriptional profiling, few have focused on the triple-negative subtype (14-17). In addition, most studies have screened genes simply based on the differentially expressed genes (DEGs) between TNBC and adjacent or normal tissues, in which the selected genes may not be specific to the triple-negative phenotype. To address this issue, we performed a study to identify TNBC-related genes (TRGs) based on the overlapping genes between the results of differential gene expression analysis and coexpression network analysis using The Cancer Genome Atlas (TCGA). Additionally, we developed a nomogram integrating the signature and traditional clinicopathological features to improve the predictive ability and for easy use in clinical practice. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1931/rc).
Methods
Data collection and processing
Both the gene expression data (HTSeq-Counts) and clinical information of breast cancer (BRCA) were downloaded from TCGA by the TCGAbiolinks package (version 2.14.1) (18). The expression data set contains a total of 1,090 primary tumors and 113 normal tissues. Here, we performed prefiltering to remove genes that had fewer than 10 reads in total. We also included our previously published cohort of 465 patients with TNBC treated at Fudan University Shanghai Cancer Center (FUSCCTNBC) (10). The RNA-sequencing data of FUSCCTNBC are available in the Sequence Read Archive (RNA-seq: SRP157974). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential gene expression analysis
All samples were classified as different subtypes of hormone receptor-positive/HER2-negative (HR+/HER2-), HR+/HER2+, HR-/HER2+, TNBC, and normal samples according to the status of ER, PR, and HER2. The baseline characteristics of the 158 TNBC patients are presented in Table S1.
Differential gene expression analysis was performed using the DESeq2 package (version 1.26.0) (19). The corresponding fold change (FC) and adjusted P value were also obtained. The DEGs were detected between TNBC and each subtype of sample with an absolute log2FC of >1 and an adjusted P value of <0.05. The overlap of the DEGs was finally determined. We also performed differential gene expression analysis between TNBC and non-TNBC samples (HR+/HER2-, HR+/HER2+, and HR-/HER2+).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using the “clusterProfiler” package (version 3.14.3) (20) and “msigdbr” package (version 7.0.1) (21). The differential gene expression analysis outputs described above in the TNBC vs. non-TNBC analysis were used to prepare the ranked gene list and 1000 permutations were used in our study.
Coexpression network construction and module identification
First, variance-stabilizing transformation was adopted to transform the count data. Variance analysis was then performed, and genes with the top 25% variance (14,128 genes) were selected for weighted gene coexpression network analysis (WGCNA).
A gene coexpression network was constructed based on the expression set of the selected genes using the WGCNA package (version 1.69) (22). We then constructed an adjacency matrix by calculating the Pearson correlation coefficient between each pair of genes. In our study, the power of β=5 (scale-free R2=0.9) was chosen as an optimal soft threshold to guarantee a scale-free network. We also calculated the topological overlap measure (TOM), which represents the overlap in shared neighbors, based on the adjacency matrix to further identify functional modules in the coexpression network.
The module eigengene (ME) was defined as the first principal component of the corresponding module and was regarded as an optimal interpretation of the gene expression profile of a given module (22). The correlation coefficient was calculated between MEs and clinical traits to identify the highly related modules. Modules with the top 3 correlations were identified.
Gene Ontology and pathway enrichment analysis
The R package “clusterProfiler” (version 3.14.3; The R Foundation for Statistical Computing, Vienna, Austria) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the genes in the selected modules (20). The ontology is categorized into 3 categories: molecular function (MF), biological process (BP), and cellular component (CC).
Gene screening and prognostic signature development
We first identified the TRGs, namely, the intersection between selected modules related to TNBC and DEGs between TNBC and the other 4 subtypes of samples. All TRGs were grouped by the median value of gene expression. The survival probability distribution of the TRGs was estimated based on the Kaplan-Meier method and compared using the log-rank test.
Least absolute shrinkage and selection operator (LASSO) Cox regression was employed with 10-fold cross-validation to further filter the prognosis-related TRGs obtained above (23). The LASSO Cox regression was performed using the “glmnet” package (version 3.0-2).
Development of the prognostic signature and nomogram
The gene signature was established according to the LASSO Cox regression coefficients described above.
Before developing the nomogram, multivariable analysis was performed to identify the spectrum of prognostic factors, and a nomogram was built by integrating the gene signature and clinicopathological parameters to predict the survival probability of different time points using the “rms” package (version 5.1-4).
Validation and performance of the model
The bootstrap resampling method with 1000 resamples was adopted for internal validation of the risk score and estimation of the confidence intervals (CIs) for point estimates of performance metrics.
Model performance was quantified and compared using Harrell’s concordance index (C-index) and Bayesian information criterion (BIC). In our study, the net reclassification index (NRI) for event probability at different time points was also adopted to assess the reclassification performance and improvement of the model (24). When the baseline and new models were nested, NRI >0 indicated improved performance of the new model. Calibration curves were estimated to assess the calibration performance of the nomogram.
To assess the performance of the gene signature as a categorical predictor, the optimal cutoff was determined using X-tile software (version 3.6.1; Yale University, New Haven, CT, USA) and multivariate analysis was also performed to evaluate the association between categorical predictors and OS.
Statistical analysis
Summary statistics and frequency tabulation were used to characterize the data distribution. The t test, the Mann-Whitney-Wilcoxon test, and the Kruskal-Wallis test were used for comparisons of continuous variables and ordered categorical variables, while Pearson chi-square test or Fisher exact test was used to compare unordered categorical variables. All tests were performed in R software (version 3.6.3; r-project.org), and a 2-sided P value of <0.05 was adopted to indicate statistical significance.
Results
DEGs of TNBC among breast cancer subtypes
The upset plot displayed the DEGs between TNBC and other subtypes of samples in TCGA data sets (Figure 1A,1B). A total of 4,417 DEGs were identified between TNBC and the HR+/HER2- subtype, 8,536 DEGs between TNBC and HR+/HER2+, 4,187 between TNBC and HR-/HER2+, and 6,847 between TNBC and normal tissues. After taking the intersection, a total of 864 overlapping DEGs were identified.
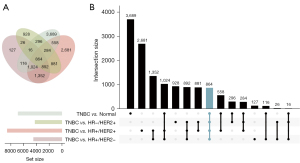
Additionally, a total of 3,969 DEGs were identified between TNBC and non-TNBC. The GSEA plots indicated that most DEGs were associated with the inflammatory response or cell cycle regulation (E2F_TARGETS and G2M CHECKPOINT) (Figure S1).
Construction of the coexpression network and identification of key modules
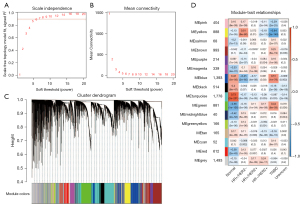
We performed WGCNA to further explore the gene modules associated with different subtypes of breast cancer, and all tumor samples were clustered based on the average linkage method and the Pearson correlation method. As stated in the Methods section, the power of β=5 (scale-free R2=0.90) was selected as the soft threshold to guarantee a scale-free network (Figure 2A,2B). A total of 16 modules were generated via hierarchical clustering, and each module was marked by various colors (Figure 2C). Inside, yellow, blue, and green modules were considered highly related to the trait of TNBC and were chosen for further analysis (Figure 2D). Both a distinctly different pattern of gene expression for TNBC (Figure S2A-S2C) and a high correlation between module membership and gene significance (Figure S2D-S2F) were observed in each of the yellow, blue, and green modules.
GO and KEGG pathway enrichment analysis
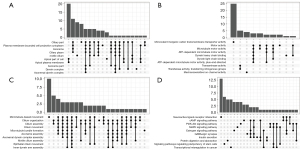
Genes in 3 modules were annotated in 4 aspects, namely CC, MF, BP, and KEGG. In the yellow module, genes were mainly enriched in ciliary part, plasma membrane bounded cell projection cytoplasm, and motile cilium in the CC category (Figure 3A). In terms of MF, genes were mostly enriched in monovalent inorganic cation transmembrane transporter activity, motor activity, and dynein heavy/light chain binding (Figure 3B). In terms of BP, genes were mainly enriched in microtubule-based movement, cilium movement, and microtubule bundle formation (Figure 3C). In the KEGG category, genes were mainly enriched in neuroactive ligand-receptor interaction and the cAMP signaling pathway, the PI3K-Akt signaling pathway, and the MAPK signaling pathway (Figure 3D). In the blue module, genes were mainly enriched in cell-cell junction, neuronal cell body, and synaptic membrane of the CC category; metal ion transmembrane transporter activity, monovalent inorganic cation transmembrane transporter activity, and serine-type peptidase activity of the MF category; epidermis development, organic anion transport, and epidermal cell differentiation of the BP category; and the PI3K-Akt signaling pathway, the cAMP signaling pathway, and the MAPK signaling pathway of the KEGG category (Figure S3). In the green module, genes were mostly enriched in chromosomal region, spindle, and condensed chromosome of the CC category; tubulin binding, ATPase activity, and microtubule binding of the MF category; organelle fission, nuclear division, and chromosome segregation of the BP category; and cell cycle, oocyte meiosis, and cellular senescence of the KEGG category (Figure S4).
Establishment and prognostic value of the signature
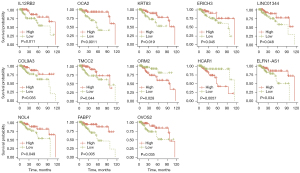
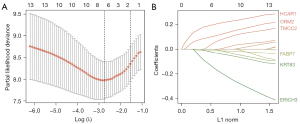
A total of 267 TRGs were determined when we took the intersection between the DEGs (TNBC vs. other 4 subtypes of samples) and genes in the yellow, blue, and green modules. Some 13 of them were associated with OS, namely COL9A3, ERICH3, ELFN1-AS1, FABP7, HCAR1, IL12RB2, KRT83, LINC01344, NOL4, OCA2, TMCC2, OVOS2, and ORM2 (Figure 4). Using the LASSO Cox regression model, 6 TRGs with nonzero coefficients were evaluated for their prognostic value (Figure 5A,5B). The following calculation formula of the prognostic signature was therefore derived: risk score =0.1944× HCAR1 +0.0999× ORM2 +0.0365×TMCC2 −0.1338× ERICH3 −0.0231× KRT83 −0.0128× FABP7.
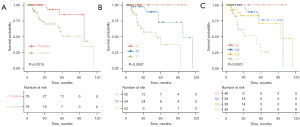
A significant difference in prognosis was observed between patients with different risk scores when the score was stratified by median (log-rank P=0.0015), tertile (log-rank P<0.0001), and quartile (log-rank P<0.0001) values (Figure 6A-6C). With a 1-unit increase in the risk score, a 4.34-fold risk of death was observed [hazard ratio (HR) 4.34; 95% CI: 2.35–8.03; P<0.001]. After adjusting for clinicopathological variables, 7.49-fold risk of death was observed (HR 7.49; 95% CI: 3.57–15.70; P<0.001; Table S2).
In addition, we also performed the analysis when the gene signature was used as a categorical predictor with a threshold of 2.5. A total of 131 (82.9%) patients were classified as low risk (<2.5), and a small proportion (17.1%) were categorized as high risk. After adjustment for age, menopausal status, race, histology, pathological stage, and breast and axillary surgery, the categorical gene signature also remained prognostic (HR 15.70; 95% CI: 3.67–67.22; P<0.001).
Establishment of the nomogram
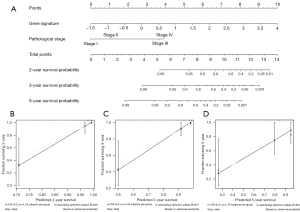
Multivariable analysis showed that both the risk score and pathological stage were independent prognostic factors in patients with TNBC (Table S2); thus, we developed a nomogram integrating the risk score and pathological stage for easy use in clinical practice to predict the 2-, 3-, and 5-year survival probability (Figure 7A), and the calibration curve showed good calibration for different timepoints (Figure 7B-7D).
Performance of the prognostic signature and nomogram
Model performance of the pathological stage, risk score, and nomogram was then assessed. The C-index increased from 0.80 for pathological stage to 0.83 for the risk score (ΔC-index 0.03; 95% CI: –0.06 to 0.11) and to 0.89 for the nomogram (ΔC-index 0.08; 95% CI: 0.01–0.14). A similar trend was observed in terms of BIC, which was decreased from 145.46 for pathological stage to 130.78 for the risk score and to 125.21 for the nomogram, indicating better performance. When compared with pathological stage, the risk score had a 2-year NRI of 0.90 (95% CI: –1.07 to 1.58), 3-year NRI of 0.51 (95% CI: –0.84 to 1.41), and 5-year NRI of 0.64 (95% CI: –0.48 to 1.46). Likewise, the nomogram had a 2-year NRI of 1.85 (95% CI: 0.01–1.14), 3-year NRI of 1.53 (95% CI: 0.30–2.10), and 5-year NRI of 1.00 (95% CI: 0.27–1.78) (Table 1).
Table 1
| Metrics | Pathological stage (95% CI) | Risk score (95% CI) | Pathological stage plus risk score (nomogram) (95% CI) |
|---|---|---|---|
| Performance of the risk score in TCGA to BRCA | |||
| C-index | 0.80 | 0.83 | 0.89 |
| BIC | 145.46 | 130.78 | 125.21 |
| Validation by bootstrap resampling method | |||
| C-index | 0.80 (0.71 to 0.89) | 0.84 (0.75 to 0.92) | 0.90 (0.81 to 0.98) |
| BIC | 141.35 (78.59 to 204.11) | 129.82 (73.21 to 186.44) | 120.95 (65.04 to 176.86) |
| Pairwise comparison of the performance | |||
| ΔC-index | Reference | 0.03 (−0.06 to 0.11) | 0.08 (0.01 to 0.14) |
| – | Reference | 0.06 (0.00 to 0.11) | |
| 2-year NRI | Reference | 0.90 (−1.07 to 1.58) | 1.85 (0.67 to 2.26) |
| – | Reference | 1.53 (0.79 to 2.02) | |
| 3-year NRI | Reference | 0.51 (−0.84 to 1.41) | 1.53 (0.30 to 2.10) |
| – | Reference | 1.12 (0.26 to 1.86) | |
| 5-year NRI | Reference | 0.64 (−0.48 to 1.46) | 1.00 (0.27 to 1.78) |
| – | Reference | 0.16 (−0.57 to 1.17) | |
The 2- , 3-, and 5-year NRI indicates the NRIs which are calculated by the event (death) probability at the 2nd, 3rd, and 5th year. C-index, concordance index; TCGA, The Cancer Genome Atlas; BRCA, breast cancer; BIC, Bayesian information criterion; NRI, net reclassification index; CI, confidence interval.
Additionally, we compared the risk score and nomogram. Similar results were observed. The C-index increased with a ΔC-index of 0.06 (95% CI: 0.00–0.11). In terms of reclassification performance, the nomogram had a 2-year NRI of 1.53 (95% CI: 0.79–2.02), 3-year NRI of 1.12 (95% CI: 0.26–1.86), and 5-year NRI of 0.16 (95% CI: –0.57 to 1.17) (Table 1). All the data presented above indicated an improved performance after the addition of the gene signature to pathological stage.
When validated by the bootstrap resampling method, the risk score alone showed a robust performance, with a C-index of 0.84 (95% CI: 0.75–0.92) and BIC of 129.82 (95% CI: 73.21–186.44); the nomogram had a C-index of 0.90 (95% CI: 0.81–0.98) and BIC of 120.95 (95% CI: 65.04–176.86) (Table 1). We performed external validation using our previously published FUSCC-TNBC data set. When used as a continuous predictor, higher risk scores suggested a worse prognosis (HR 2.9; 95% CI: 1.3–6.4; P=0.0071). A significant difference in prognosis was shown among individuals with various risk scores when stratified by the top quartile values (log-rank P=0.0039; Figure S5). Notably, the model of the pathological stage combined with risk score also exhibited a good performance (C-index =0.72).
Discussion
Our study mainly focused on the association between genomics and clinical outcomes in patients with TNBC. We established a risk score based on the gene expression profiling of TRGs, which were obtained from the intersection between TNBC-specific modules in WGCNA and DEGs (TNBC vs. the other 4 subtypes). With a 1-unit increase in the risk score, an approximately 7-fold higher risk of death was observed in the current study.
According to various classifications, such as gene expression, metabolic pathways, or proteomic data, TNBC can be further subdivided into diverse subtypes. Each of these subtypes presents its own distinct features. Ensenyat-Mendez et al. reported that different classifications can be based on gene expression, metabolic pathways, methylation, and other means (11). Vasudevan et al. classified TNBCs into at least 17 different subtypes via proteomic analysis (12). Moreover, Dawson et al. identified at least 10 subtypes based on genomic and transcriptomic landscapes (13). Therefore, TNBC is known to be a highly heterogeneous disease with variable prognosis. Traditional clinicopathological markers, such as grade, tumor size, and lymph node status, are routinely adopted for prognosis prediction in the management of breast cancer. However, the prognostic value of these parameters remains questionable and only a marginal association has been observed between conventional markers and prognosis in TNBC (4,25,26). All these studies reinforce the idea that conventional biomarkers alone cannot yield accurate prognosis prediction. Additionally, the identification of genomic markers that may inform prognosis and guide treatment decisions for TNBC remains a clinically unmet need. To address this issue, a nomogram was established in our study by integrating the genomic signature and pathological stage. Improved performance was found when compared with the gene signature or pathological stage alone.
The GO and KEGG pathway analyses revealed the TNBC-related enriched pathways. Dong et al. reported that activation of the cAMP signaling system inhibits the migration and motility of TNBC cells (27). Many reports have shown that the PI3K-Akt signaling pathway and the MAPK signaling pathway play an important role in TNBC (28-31). The development of medications targeting the PI3K/Akt signaling pathway or the MAPK signaling pathway for the treating of TNBC is a developing sector that must consider the efficacies and toxicity of novel therapies as well as their interactions with other cancer pathways. Interestingly, Zhang et al. reported that estrogen signaling is associated with the progression of ER-negative breast cancer (32). However, the GABAergic synapse, protein digestion and absorption, neuroactive ligand-receptor interaction pathways, and others, have not been thoroughly studied in TNBC. These pathways may be involved in the development of TNBC. Therefore, additional studies are needed to discover whether targeting these pathways has therapeutic promise.
Prognostic signatures derived from high-dimensional data may suffer from overfitting to a certain extent. To address this problem, a Cox regression model with a LASSO penalty has been applied for the selection of genes and the shrinkage of corresponding coefficients; this can facilitate the selection of genes with high expression variance and avoid multicollinearity, thus contributing to improved prognostic performance (23).
Several of the 6 genes involved in the signature have been reported to play potential roles in cancer biology. For example, HCAR1 has been reported to act as a sensor of extracellular lactate in cancer (33). Activation of HCAR1 may contribute to increased expression of monocarboxylate transporters, which are crucial for lactate uptake as an alternative energy source for cancer cells (33-35). It may also mediate resistance to anticancer drugs in cervical cancer (34,35). ORM2, an acute phase plasma protein, may be involved in aspects of immunosuppression (36). The plasma ORM2 concentration was found to be increased in breast and ovarian cancers and was also reported to be an independent prognostic factor for colorectal cancer (37,38). Further, FABP7 has been widely studied and was found to be mainly expressed in the nervous system and mammary gland. In the mammary gland, FABP7 promotes differentiation and suppresses the proliferation of mammary cells (39). Kwong et al. also reported the antitumorigenic function of FABP7 in TNBC by activating the expression of PPAR-α and therefore limiting the efficient utilization of available bioenergetic substrates, such as glucose, and impacting metabolic plasticity (40). In addition, both Zhang et al. and Alshareeda et al. reported better prognosis in FABP7-positive triple-negative disease (41,42). Consistently, our study found that increased expression of FABP7 acted as a protective factor in patients with TNBC.
To date, several gene signatures have been proposed to inform prognosis for patients with TNBC. Jiang et al. developed an integrated messenger RNA (mRNA)–long noncoding RNA (lncRNA) signature based on transcriptome analysis for 33 paired TNBC and adjacent normal breast tissues (14). Yang et al. identified several mRNAs, lncRNAs, and micro RNAs (miRNAs) that may serve as potential biomarkers for prognosis prediction in TNBC patients based on the differential expression analysis between 111 TNBC tissues and 104 noncancerous tissues (17). As stated before, most previous studies have selected genes that were differentially expressed between TNBC and normal samples. Nevertheless, these screened genes may not be specific to TNBC’s expression profiling nor to the other subtypes. In the present study, differential gene expression analyses were performed between TNBC and HR+/HER2-, HR+/HER2+, HR-/HER2+, and normal tissues. Coexpression network analysis was also conducted to identify TNBC-related gene modules. However, cautious interpretation about TNBC-related genes should be noted since no experimental validation was performed in our study. Finally, the genes in the intersection between the DEGs and the selected TNBC-related modules were named TNBC-related genes and were adopted for signature development.
Inevitably, this work had several limitations. First, we used a retrospective study design, and therefore, the prognostic signature requires rigorous validation in prospective trials. Second, the potential role of several genes involved in the signature in TNBC biology has not been thoroughly identified, and thus, further experimental studies are required to facilitate our understanding of these genes.
Conclusions
The mRNA-based prognostic signature and the nomogram reported herein, which incorporate both the clinicopathological parameters and gene expression profiles in the transcriptome, may inform the prognosis of patients with TNBC. Further validation in prospective settings may contribute to model generalization, facilitate counseling, and provide additional information when determining treatment approaches.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1931/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1931/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1931/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1931/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA 2019;321:288-300. [Crossref] [PubMed]
- Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017;389:2430-42. [Crossref] [PubMed]
- Kumar P, Aggarwal R. An overview of triple-negative breast cancer. Arch Gynecol Obstet 2016;293:247-69. [Crossref] [PubMed]
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429-34. [Crossref] [PubMed]
- Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7:683-92. [Crossref] [PubMed]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48. [Crossref] [PubMed]
- Garrido-Castro AC, Lin NU, Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov 2019;9:176-98. [Crossref] [PubMed]
- Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67. [Crossref] [PubMed]
- Jiang YZ, Ma D, Suo C, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019;35:428-440.e5. [Crossref] [PubMed]
- Ensenyat-Mendez M, Llinàs-Arias P, Orozco JIJ, et al. Current Triple-Negative Breast Cancer Subtypes: Dissecting the Most Aggressive Form of Breast Cancer. Front Oncol 2021;11:681476. [Crossref] [PubMed]
- Vasudevan S, Adejumobi IA, Alkhatib H, et al. Drug-Induced Resistance and Phenotypic Switch in Triple-Negative Breast Cancer Can Be Controlled via Resolution and Targeting of Individualized Signaling Signatures. Cancers (Basel) 2021;13:5009. [Crossref] [PubMed]
- Dawson SJ, Rueda OM, Aparicio S, et al. A new genome-driven integrated classification of breast cancer and its implications. EMBO J 2013;32:617-28. [Crossref] [PubMed]
- Jiang YZ, Liu YR, Xu XE, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res 2016;76:2105-14. [Crossref] [PubMed]
- Pece S, Disalvatore D, Tosoni D, et al. Identification and clinical validation of a multigene assay that interrogates the biology of cancer stem cells and predicts metastasis in breast cancer: A retrospective consecutive study. EBioMedicine 2019;42:352-62. [Crossref] [PubMed]
- Wang K, Li HL, Xiong YF, et al. Development and validation of nomograms integrating immune-related genomic signatures with clinicopathologic features to improve prognosis and predictive value of triple-negative breast cancer: A gene expression-based retrospective study. Cancer Med 2019;8:686-700. [Crossref] [PubMed]
- Yang R, Xing L, Wang M, et al. Comprehensive Analysis of Differentially Expressed Profiles of lncRNAs/mRNAs and miRNAs with Associated ceRNA Networks in Triple-Negative Breast Cancer. Cell Physiol Biochem 2018;50:473-88. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739-40. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) 1996;58:267-88. [Crossref]
- Uno H, Tian L, Cai T, et al. A unified inference procedure for a class of measures to assess improvement in risk prediction systems with survival data. Stat Med 2013;32:2430-42. [Crossref] [PubMed]
- Wen S, Manuel L, Doolan M, et al. Effect of Clinical and Treatment Factors on Survival Outcomes of Triple Negative Breast Cancer Patients. Breast Cancer (Dove Med Press) 2020;12:27-35. [Crossref] [PubMed]
- Bae MS, Moon HG, Han W, et al. Early Stage Triple-Negative Breast Cancer: Imaging and Clinical-Pathologic Factors Associated with Recurrence. Radiology 2016;278:356-64. [Crossref] [PubMed]
- Dong H, Claffey KP, Brocke S, et al. Inhibition of breast cancer cell migration by activation of cAMP signaling. Breast Cancer Res Treat 2015;152:17-28. [Crossref] [PubMed]
- Costa RLB, Han HS, Gradishar WJ. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res Treat 2018;169:397-406. [Crossref] [PubMed]
- Khan MA, Jain VK, Rizwanullah M, et al. PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: a review on drug discovery and future challenges. Drug Discov Today 2019;24:2181-91. [Crossref] [PubMed]
- Wang L, Zhou Y, Jiang L, et al. CircWAC induces chemotherapeutic resistance in triple-negative breast cancer by targeting miR-142, upregulating WWP1 and activating the PI3K/AKT pathway. Mol Cancer 2021;20:43. [Crossref] [PubMed]
- Chen MS, Yeh HT, Li YZ, et al. Flavopereirine Inhibits Autophagy via the AKT/p38 MAPK Signaling Pathway in MDA-MB-231 Cells. Int J Mol Sci 2020;21:5362. [Crossref] [PubMed]
- Zhang XT, Kang LG, Ding L, et al. A positive feedback loop of ER-α36/EGFR promotes malignant growth of ER-negative breast cancer cells. Oncogene 2011;30:770-80. [Crossref] [PubMed]
- Roland CL, Arumugam T, Deng D, et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res 2014;74:5301-10. [Crossref] [PubMed]
- Wagner W, Ciszewski WM, Kania KD. L- and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun Signal 2015;13:36. [Crossref] [PubMed]
- Wagner W, Kania KD, Blauz A, et al. The lactate receptor (HCAR1/GPR81) contributes to doxorubicin chemoresistance via ABCB1 transporter up-regulation in human cervical cancer HeLa cells. J Physiol Pharmacol 2017;68:555-64. [PubMed]
- Fayazfar S, Zali H, Arefi Oskouie A, et al. Early diagnosis of colorectal cancer via plasma proteomic analysis of CRC and advanced adenomatous polyp. Gastroenterol Hepatol Bed Bench 2019;12:328-39. [PubMed]
- Gao F, Zhang X, Whang S, et al. Prognostic impact of plasma ORM2 levels in patients with stage II colorectal cancer. Ann Clin Lab Sci 2014;44:388-93. [PubMed]
- Duché JC, Urien S, Simon N, et al. Expression of the genetic variants of human alpha-1-acid glycoprotein in cancer. Clin Biochem 2000;33:197-202. [Crossref] [PubMed]
- Yang Y, Spitzer E, Kenney N, et al. Members of the fatty acid binding protein family are differentiation factors for the mammary gland. J Cell Biol 1994;127:1097-109. [Crossref] [PubMed]
- Kwong SC, Jamil AHA, Rhodes A, et al. Metabolic role of fatty acid binding protein 7 in mediating triple-negative breast cancer cell death via PPAR-α signaling. J Lipid Res 2019;60:1807-17. [Crossref] [PubMed]
- Alshareeda AT, Rakha EA, Nolan CC, et al. Fatty acid binding protein 7 expression and its sub-cellular localization in breast cancer. Breast Cancer Res Treat 2012;134:519-29. [Crossref] [PubMed]
- Zhang H, Rakha EA, Ball GR, et al. The proteins FABP7 and OATP2 are associated with the basal phenotype and patient outcome in human breast cancer. Breast Cancer Res Treat 2010;121:41-51. [Crossref] [PubMed]
(English Language Editors: J. Jones and J. Gray)


