Machine learning improves risk stratification of coronary heart disease and stroke
Introduction
Cardiovascular diseases (CVDs) are a leading cause of global mortality and morbidity. In China, 93.8 million CVD patients were reported in 2016, more than twice the number reported in 1990 (40.6 million) (1). By 2019, cardiovascular death accounted for 45.19% of total deaths in rural areas and 43.56% in urban areas. The incessant increase in CVD prevalence has been highlighted as a global health challenge (2). Therefore, improving the efficiency of the CVD healthcare system is an urgent task.
It is now widely accepted that machine learning can improve medical efficiency by assisting doctors in analyzing large-scale high-dimensional clinical data. The merit of machine learning has been consistently supported by several predictive and risk stratification studies (3), including those for CVD and its subtypes (4,5). Notably, an effective stroke classification model was trained using a Chinese prospective cohort with 56 stroke patients and 1,075 non-stroke participants. This study applied a synthetic minority over-sampling technique (SMOTE) method for data balancing, which achieved an area under the receiver operating characteristic curve (AUC) of 0.72 (6). It should be noted that CVD patients appear relatively rarely in the natural population, which highlights the potential imbalance issues in model training. Early risk assessment may prevent cardiovascular events in the future (7). In clinical practice, people often have poor compliance (8) and it may be difficult to identify people at high risk of CVD. Therefore, an efficient risk stratification model is necessary. Furthermore, the Asian population is underrepresented in published studies. As illustrated in Liu’s research, the models based on the European population may overestimate the risk in Asian populations (9). To address the problems of imbalance and population difference, we integrated the machine learning models with the under-sampling method to create a coronary heart disease (CHD) and cerebral ischemic stroke (CIS) risk stratification model that is clinically applicable for the Asian population. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1916/rc).
Methods
Study design
A case-control study was conducted with participants attending a physical examination at Tongji Hospital in Wuhan, China, from May 2008 to December 2019. The inclusion criteria were as follows: (I) patients aged 30 years and older; and (II) patients without a history of tumor, liver cirrhosis, or renal failure. Participants with a recorded CHD or CIS history were considered cases. Participants without any history of CHD and CIS were considered controls.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee at Tongji Medical College, Huazhong University of Science and Technology (No. TJ-IRB20191215). Individual consent for this retrospective analysis was waived.
Clinical indicators and inclusion criteria
The medical records of all the participants were collated at the Tongji Hospital during standardized in-person interviews (10). A total of 118 indicators were selected from medical records, including lifestyle characteristics and clinical measurements. The standardized protocols of data collection can be found in the Supplementary Methods (Appendix 1).
The diagnosis of CIS was based on symptoms and cerebral infarction confirmed by computed tomography or magnetic resonance imaging; CHD was diagnosed according to symptoms (mainly angina) and electrocardiography or coronary angiography. Patients with self-reported CHD, coronary artery bypass grafting, coronary stent implantation, percutaneous coronary intervention, or percutaneous transluminal coronary angioplasty were also considered CHD patients. The patient’s self-reports were confirmed by doctors through the medical insurance system.
Imputation
Indicators with a missing rate greater than 45% were discarded, and the remaining missing indicators were imputed (Table S1). Random forest was applied to impute the missing continuous data with the R package missForest v1.4 (11). Missing categorical data were imputed by the median of each feature.
Model establishment
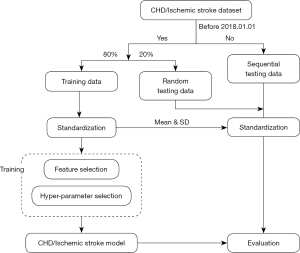
CHD and CIS datasets were built to train the CHD and CIS models. The unreliable data were excluded, defined by inconsistency in gender, age (differences larger than 5 years), or physical examination time with cases.
In each dataset, 80% of the data before 1 January 2018 were randomly sampled as the training set, and the remaining 20% were used for random testing. The data collected after the year 2018 were used for sequential testing.
All continuous variables were standardized with a mean of 0 and a variance of 1. Three feature selection approaches were applied for continuous features: (I) analysis of variance (ANOVA) (12); (II) recursive feature elimination (RFE) (13); and (III) Boruta (14). The P value threshold of ANOVA was set at 0.05. The basic estimator of both RFE and Boruta was a random forest model with default parameters (15). Continuous features significant in at least two methods were selected for subsequent analysis.
For categorical features, the VarianceThreshold method was used to exclude the features that were either 1 or 0 in more than 80% of the samples. All feature selection methods were performed in Python 3.7 using the packages sklearn v0.24 and Boruta v0.3 (14,16).
A bagging classifier was developed with additional balancing from BalancedBaggingClassifier using the python package scikit-multilearn v0.2 (17). Specifically, the base estimator was the random forest model (15). Random down-sampling without replacement was used to produce a more balanced dataset. The ratio of major classes to minor classes was 1, and the number of downsampling (number of base models) was 100. The training process included hyper-parameters selection and model simplification. The hyper-parameters (n_estimators, max_depth, and min_samples_split) were searched to maximize the AUC score using GridSearchCV in 5-fold cross-validation. To improve the efficiency in clinical practice, an approach was applied to reduce the number of indicators while ensuring the utility of the risk stratification model. The Shapley Additive exPlanations (SHAP) explainer (18) was constructed to rank the features using Python package shap v0.39, and 1,000 randomly selected samples were used to calculate the feature contribution. We used 80% of the randomly selected sets to train a model, and the remaining 20% of the samples were used for model evaluation. The features with a higher ranking were included unless no improvements were observed. The above training and validation process was repeated 100 times to secure robustness. Measures (average precision, specificity, sensitivity, F-score, and AUC) were used to systematically evaluate the performance. The trained model was interpreted by SHAP.
To better evaluate the performance of the final model, the complex model before reducing feature numbers, random forest model, logistic regression model, and support vector machine model was constructed. Constructions of the above three traditional machine learning models used the same characteristics as the complex model. Framingham risk scores (FHS) of CHD and CIS were calculated. The risk prediction equations of CHD (19) and CIS (20) for 10 years are presented in Supplementary Methods.
Development of risk stratification Python package
A Python package named CCRS (coronary heart disease and cerebral ischemic stroke risk stratification) was developed for generating better application in clinics by providing population risk and relative risk. Population risk would be categorized into mild, moderate, and severe. In detail, based on CHD and CIS models’ prediction in the training data, the threshold between mild and moderate corresponded to the true positive rate of 95%, and the threshold between severe and moderate corresponded to the true negative rate of 95%. The relative risk would be presented as risk ranking among peers. For the purpose of clarity, with the individual predicted value and age, the ranking of the individuals in the same age group of the training set was provided as relative risk.
Statistical analysis
Continuous variables were presented as means and standard deviations. Categorical variables were described as counts and percentages. To evaluate the difference between case and control groups, continuous indicators were analyzed by the rank sum test and categorical indicators were analyzed by the chi-square test. Delong methods were used to test the differences in the AUCs of the receiver operating characteristic (ROC) curves.
Results
Basic characteristics of the study population
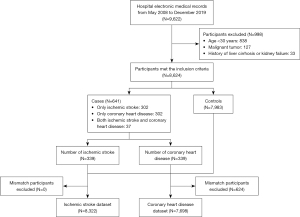
This study included 7,983 (92.6%) controls and 641 (7.4%) cases (Figure 1). Among the cases, 302 were CHD patients and 302 were CIS patients, and 37 people had both CHD and CIS. The mean age of the study population was 51±11.4 years and 71.3% of the study population were males.
All 118 clinical indicators are presented in Table S2. The lifestyle characteristics and laboratory results relevant to blood pressure, glucose, and lipids are summarized in Table 1. The proportion of smokers and drinkers in the study population was 32.2% and 44.3%, respectively. In terms of vascular stiffness, 99.2% of the population had normal ankle-brachial index (ABI; 0.9≤ ABI <1.4) (21), and 55.7% of the population had normal brachial-ankle pulse wave velocity (baPWV <1,400) (22). These characteristics in the CHD dataset (N=7,698) and CIS dataset (N=8,322) are presented in Table S3 and Table S4.
Table 1
| Variables | Population (N=8,624) |
|---|---|
| Basic characteristics | |
| Age, years | 51.9±11.4 |
| Sex | |
| Male | 6,152 (71.3) |
| Female | 2,472 (28.7) |
| Smoker | 2,777 (32.2) |
| Drinker | 3,823 (44.3) |
| BMI, kg/m2 | 24.6±3.1 |
| Cardiovascular medical history | |
| CHD | 339 (3.9) |
| CIS | 339 (3.9) |
| Clinical measurements | |
| SBP, mmHg | 126.3±16.9 |
| DBP, mmHg | 81.7±11.4 |
| LDL-C, mmol/L | 2.9±0.8 |
| HDL-C, mmol/L | 1.2±0.3 |
| TC, mmol/L | 4.7±0.9 |
| TG, mmol/L | 1.6±1.4 |
| FBG, mmol/L | 5.2±1.2 |
| HbA1c, % | 5.8±0.7 |
| baPWV, cm/s | 1,409.7±256.5 |
| <1,400 | 4,800 (55.7) |
| 1,400–1,800 | 3,191 (37.0) |
| >1,800 | 633 (7.3) |
| ABI | 1.1±0.07 |
| <0.9 | 59 (0.7) |
| 0.9–1.4 | 8,554 (99.2) |
| >1.4 | 11 (0.1) |
Continuous variables are described as mean ± SD. Categorical variables are described as number (percentage). BMI, body mass index; CHD, coronary heart disease; CIS, cerebral ischemic stroke; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; FBG, fasting blood glucose; HbA1c, hemoglobin A1C; baPWV, brachial-ankle pulse wave velocity; ABI, ankle-brachial index; SD, standard deviation.
The ensemble learning model effectively distinguished CHD patients and CIS patients
The flowchart of model development is shown in Figure 2. After standardization, the first step of training was the elimination of irrelevant and redundant information. In this step, VarianceThreshold, ANOVA, RFE, and Boruta were used for features selection. After selection, only 5 of the 55 discrete variables presented significant contribution in both CHD and CIS patients, namely, sex, smoking status, drinking, vascular stiffness, and hypertension. Meanwhile, 23 of the 59 continuous variables were considered important in the CHD dataset, and 22 of the 59 variables showed importance in the CIS dataset (Table S4). Five features, namely, age, prostate-specific antigen (PSA), total cholesterol (TC), baPWV, and estimated glomerular filtration rate (eGFR), were considered crucial in both CHD and CIS patients (Figure S1, Table S5).
GridSearchCV analysis was then applied to optimize the balanced bagging classifier (BBC) model. The optimal hyper-parameters for the optimal CHD classification performance were as follows: estimators 100, maximum depth 6, and minimum sample split 7 (Figure S2). In terms of the CIS model, the ideal hyper-parameters were as follows: estimators 500, maximum depth 4, and minimum sample split 2 (Figure S3).
We then used SHAP analysis to reduce the model complexity. The CHD model performance was retained when including the top 8 features (Figure S4). For the CIS model, only 5 features were required for maintaining the model efficiency (Figure S5). Lastly, the final ensemble models were built based on the optimized parameters and features.
To evaluate the model performance, multiple reported models were constructed, including Framingham risk score (19,20), BBC model before reducing feature numbers, and traditional machine learning models (logistic regression, support vector machine, and random forest), by applying multiple measures. These measures included ROC curves, sensitivity, specificity, F score, and average precision. In the CHD model, the AUCs of the random and the sequential validation were 0.895 and 0.905, respectively (Figure 3A). The area under the precision-recall curves (AUPRCs) of the random and the sequential validation was 0.360 and 0.304, respectively (Figure 3B). The sensitivity of the random and the sequential validation was 0.807 and 0.873, and the specificity was 0.831 and 0.788, respectively (Table 2). Compared with other models horizontally, the model with the best performance was the BBC model before simplification, and the final BBC model performed similarly to that of the complex BBC model in the sequential test set (Figure S6). In the CIS model, the AUCs of the random and the sequential test set were 0.884 and 0.889, respectively (Figure 3C). The AUPRCs of the random and the sequential validation were 0.298 and 0.216, respectively (Figure 3D). The sensitivity of the random and the sequential validation was 0.808 and 0.877, and the specificity was 0.750 and 0.756, respectively (Table 3). Interestingly, the performance of the final model was superior to that of other CIS models horizontally (Figure S7). Notably, in both models, the predictive performance of the sequential validation was better than those of the random validation. Considering that the AUC can only reflect the discrimination of the models, the calibration of the models was evaluated (Figure S8). Unfortunately, the calibration of the models was generally poor and the BBC models for CHD and CIS might overestimate the risk.
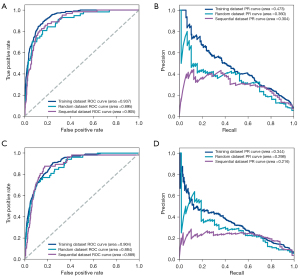
Table 2
| Models | AUC (95% CI) | Specificity | Sensitivity | F-score | AP |
|---|---|---|---|---|---|
| Random testing | |||||
| FHS-CHD | 0.708 (0.65–0.762) | 0.545 | 0.737 | 0.128 | 0.063 |
| LR | 0.901 (0.868–0.933) | 0.836 | 0.807* | 0.303 | 0.159 |
| SVM | 0.874 (0.829–0.915) | 0.829 | 0.772 | 0.283 | 0.144 |
| RF | 0.899 (0.866–0.929) | 0.815 | 0.789 | 0.274 | 0.140 |
| Complex BBC | 0.912 (0.884–0.937)* | 0.868* | 0.789 | 0.341* | 0.181* |
| BBC | 0.895 (0.860–0.928) | 0.831 | 0.807* | 0.297 | 0.155 |
| Sequential testing | |||||
| FHS-CHD | 0.73 (0.683–0.779) | 0.570 | 0.794 | 0.157 | 0.080 |
| LR | 0.900 (0.870–0.927) | 0.807 | 0.857 | 0.308 | 0.168 |
| SVM | 0.877 (0.841–0.909) | 0.806 | 0.794 | 0.287 | 0.149 |
| RF | 0.903 (0.872–0.932) | 0.774 | 0.905* | 0.289 | 0.160 |
| Complex BBC | 0.905 (0.876–0.931)* | 0.827* | 0.841 | 0.325* | 0.177* |
| BBC | 0.905 (0.877–0.931)* | 0.788 | 0.873 | 0.293 | 0.160 |
*, the highest scores in random testing or sequential testing. CHD, coronary heart disease; AUC, the area under the receiver operating characteristic curve; CI, confidence interval; AP, average precision; FHS-CHD, Framingham risk score of coronary heart disease; LR, logistic regression; SVM, support vector machine; RF, random forest; BBC, balanced bagging classifier.
Table 3
| Models | AUC (95% CI) | Specificity | Sensitivity | F-score | AP |
|---|---|---|---|---|---|
| Random testing | |||||
| FHS-CIS | 0.804 (0.757–0.851) | 0.791 | 0.596 | 0.172 | 0.075 |
| LR | 0.882 (0.848–0.913) | 0.748 | 0.885* | 0.213 | 0.112 |
| SVM | 0.839 (0.793–0.883) | 0.776 | 0.808 | 0.215 | 0.107 |
| RF | 0.865 (0.821–0.909) | 0.842* | 0.750 | 0.260* | 0.127* |
| Complex BBC | 0.880 (0.843–0.916) | 0.780 | 0.808 | 0.218 | 0.109 |
| BBC | 0.884 (0.850–0.919)* | 0.750 | 0.808 | 0.198 | 0.098 |
| Sequential testing | |||||
| FHS-CIS | 0.850 (0.812–0.886) | 0.815 | 0.719 | 0.232 | 0.110 |
| LR | 0.883 (0.847–0.912) | 0.779 | 0.825 | 0.229 | 0.117 |
| SVM | 0.813 (0.762–0.857) | 0.784 | 0.754 | 0.215 | 0.104 |
| RF | 0.872 (0.827–0.912) | 0.827* | 0.807 | 0.268* | 0.137* |
| Complex BBC | 0.877 (0.836–0.914) | 0.787 | 0.877* | 0.249 | 0.132 |
| BBC | 0.889 (0.851–0.919)* | 0.756 | 0.877* | 0.225 | 0.118 |
*, the highest scores in random testing or sequential testing. CIS, cerebral ischemic stroke; AUC, the area under the receiver operating characteristic curve; CI, confidence interval; AP, average precision; FHS-CIS, Framingham risk score of cerebral ischemic stroke; LR, logistic regression; SVM, support vector machine; RF, random forest; BBC, balanced bagging classifier.
Model interpretation
We applied SHAP analysis to interpret and understand the mechanism underlying the trained model. Our results indicated that hypertension, increased age, baPWV, hemoglobin A1c (HbA1c), and carcinoembryonic antigen (CEA) were associated with a higher risk of CHD. Meanwhile, high levels of TC, low-density lipoprotein cholesterol (LDL-C), and platelet (PLT) were associated with a lower risk of CHD (Figure 4A,4B). An increase in LDL-C was negatively correlated with the risk of CIS, and hypertension, increased age, baPWV, and ABI were positively correlated with the risk of CIS (Figure 4C,4D). In both diseases, hypertension appeared to be a common risk factor. In addition, the effects of continuous variables were linear (Spearman correlation, P<0.001, Figures S9,S10). Intriguingly, in the CHD model, the impact of CEA failed to enhance when the value was greater than 3.14. In the CIS model, the impact of ABI steeply increased when its value was greater than 1.1 (Figure 4E). In addition, the effect of age presented an S-curve in both models. To further explore the characteristics of the CEA influence curve, interaction analysis was used. The results showed that the SHAP value of CEA in individuals with high levels of LDL-C was higher in the plateau stage (Figure 4F).
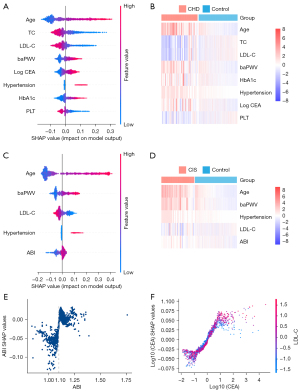
Although the feature selection process excluded the feature of gender, gender differences have been observed in CVD (23) We suspected that the effects of some other features may vary in different gender groups, which was supported by the sex-specific sub-analysis, where hypertension remained an important predictor of CHD and CIS in males, but the importance decreased in females. Other features such as age and baPWV appeared to be critical predictors in both males and females (Figure S11).
Inflammation status could be essential in CVD development
For in-depth study, the misclassifications in our model predicted accurate samples and predicted wrong controls (PWCs) were used for error analysis. The results demonstrated that the PWCs were similar to the predicted accurate patients (PAP; P>0.05), but different from the predicted accurate controls (PACs) in age, baPWV, HbA1c, CEA, and PLT (Figure S12A-S12E, P<0.001).
However, the percentage of lymphocytes in PWCs (33.0±7.9) was close to that in PACs (33.5±7.3, P=0.27) and was higher than that in PAP samples (27.2±7.4, P=0.01, Figure S12F). Considering that leukocytes are related to immunity and inflammation, we further analyzed the neutrophil count, neutrophil/lymphocyte ratio (NLR), and systemic immune inflammation index (SII) of PWCs without antiplatelet medication (Table 4). The SII in PWCs (361.9±170.4) was not only lower than that in PAPs (525.1±305.0, P<0.001), but also lower than that found in PACs (420.8±189.9, P<0.001, Table 5, Figure S13). The error analysis revealed that the PWCs were comparable to the PAPs in all 5 indicators, but different from the PACs (P<0.001) in the CIS model. Unfortunately, error analysis could not be performed in predicted wrong cases due to the limited sample size.
Table 4
| Use of antiplatelet drugs | Predicted accurate cases | Predicted accurate controls | Predicted wrong controls |
|---|---|---|---|
| Yes | 33 | 8 | 14 |
| No | 21 | 987 | 206 |
CHD, coronary heart disease.
Table 5
| Inflammatory indicators | PAC vs. PWC | PAP vs. PWC | PAC vs. PAP |
|---|---|---|---|
| Neutrophils | 0.602 | 2.75×10−2 | 3.61×10−2 |
| NLR | 0.711 | 4.30×10−3 | 1.40×10−3 |
| SII | 7.27×10−6 | 5.35×10−3 | 0.118 |
CHD, coronary heart disease; PAC, predicted accurate controls; PWC, predicted wrong controls; PAP, predicted accurate patients; NLR, neutrophil/lymphocyte ratio; SII, systemic immune inflammation index, SII was calculated by (N×P)/L (N, P, and L represent neutrophil counts, platelet counts, and lymphocyte counts, respectively).
The constructed model achieved precise risk stratification in CVD
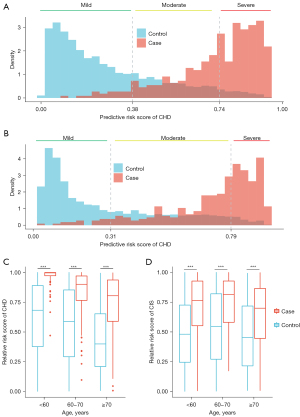
When applying the risk stratification model to the sequential testing dataset, 93.8% of CHD patients and 95.0% of CIS patients were classified into the moderate or severe group (Figure 5A,5B). The risk stratification model was used to calculate the relative risk of participants in each age group. The relative risk of CHD and CIS patients was higher than that in controls for each age group (one-sided rank-sum test P<0.001, Figure 5C,5D).
Discussion
In this study, an effective model for risk stratification in CHD and CIS was developed based on 8,624 electronic medical records among the Chinese population. With the increasing prevalence of CHD and CIS, and especially the rising incidence of CVD in young individuals (24), this tool may help individuals to understand their risk of CHD and CIS. The relative risk score provided by the model could raise awareness of early vascular aging among young individuals, to motivate them to change lifestyles following medical advice. The effectiveness of this model originates from the underlying well-performing models. When testing the performance in the sequential dataset, the AUC of CHD and CIS reached 0.905 and 0.889, respectively. Contrary to previous research (4-6), the good performance of our models may be attributed to the resolution of the imbalanced classification between controls and cases with a combination of the random downsampling and ensemble learning to ensure the maximal usage of the controls. This combination can address the data ambiguity generated from random sampling.
The relationships between CVD diseases and various indicators were also illustrated in this study. These results were consistent with our hypothesis, that the significantly related indicators of CHD and CIS were highly overlapped with each other, and only a few indicators were different. Specifically, age, baPWV, hypertension, and LDL-C were considered risk factors in both models. However, HbA1c, CEA, PLT were only found to contribute to risk in the CHD model, and ABI was only found to be crucial in the CIS model. Both ABI (25) and baPWV, as indicators of vascular stiffness, have been gradually acknowledged and applied in clinical practice. Hyperlipidemia, hyperglycemia, and hypertension are recognized as canonical risk factors for CVD (26). In this perspective, our research demonstrated that hypertension is a common risk factor in CHD and CIS and it may have a more significant effect in males than in females. This phenomenon might be attributed to the gender-specific differences in cell senescence pathways and mitochondrial function. Such differences would lead to different levels of hypertension-induced organ damage in different genders (27). Surprisingly, a negative relationship between LDL-C and disease risk was noted in both models, and also with TC in the CHD model. This may be partly due to the higher proportion of patients taking lipid-lowering drugs (Tables S3,S4). Sachdeva et al. also witnessed a negative correlation between LDL-C and CHD risk in the European population, suggesting that it might be caused by the shifts in the prevalence of other cardiovascular risk factors (28). Furthermore, it is well-known that a majority of CHD patients experience chronic low-grade inflammation (29). Consistent with previous studies (30,31), we identified CEA, one of the inflammatory indicators (32), as a potential biomarker of CHD. A total of 7.67% of patients presented with abnormally high CEA values (CEA greater than 0.5 ng/mL), compared to 3.13% of controls with abnormal values (chi-square test P<0.001). Notably, a portion of control individuals without CHD showed similarities in indicators such as age, baPWV, and HbA1c, as CHD patients, but controls had lower inflammatory levels. CVD is a complex disease, but the indicators contained in the traditional model [such as age, smoking status, and systolic blood pressure (SBP)] can only reflect part of the body’s condition. As incorporating non-traditional factors can improve the effectiveness of the model (33), adding indicators that reflect the immune system may be beneficial. Unlike SII, CEA is not in the routine set of tests, and the clinical value of this tumor marker warrants further large-scale research.
There were some potential limitations to this study. Calibration analysis showed that our models might overestimate the risk score, that is, the low-risk population might be classified as medium-to-high risk. However, such overestimation would not affect the relative risk, because the relative risk is based on ranking. In addition, it should be noted that this study only included specific medical records from an Asian population. Previous studies have suggested that the genome and the metagenome of gut microbiota may also affect the risk of CVD (34,35). The inclusion of this information may provide a better understanding of CVD risk among the population. Therefore, we are currently conducting a 10-year prospective cohort study including genome and metagenome sequencing data of all the participants in China (ChiCTR2100042724).
Conclusions
Herein, we developed 2 well-performing machine learning models for predicting CHD and CIS. The relationships between the clinical indicators and diseases were consistent with previous studies. Based on our models, we constructed a CHD and CIS risk stratification tool for the Asian population.
Acknowledgments
We would like to thank the general physicians, especially those from the Department of Geriatrics of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology for their dedication, commitment, and contribution.
Funding: This work was supported by the National Key Research and Development Program of China (No. 2020YFC2008002; principal investigator CZ) and the Major Technology Innovation of Hubei Province (No. 2019ACA141). The funding sources declare that they had no involvement in the process of this study or manuscript preparation. The contents do not reflect the official views of the government of the Hubei province.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1916/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1916/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1916/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1916/coif). All authors declare that this study was supported by the National Key Research and Development Program of China (No. 2020YFC2008002; principal investigator CZ) and the Major Technology Innovation of Hubei Province (No. 2019ACA141). BC, LY, YL, XJ, YB, and TL are employees of BGI-Shenzhen. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee at Tongji Medical College, Huazhong University of Science and Technology (No. TJ-IRB20191215) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu S, Li Y, Zeng X, et al. Burden of Cardiovascular Diseases in China, 1990-2016: Findings From the 2016 Global Burden of Disease Study. JAMA Cardiol 2019;4:342-52. [Crossref] [PubMed]
- The Writing Committee of the Report on Cardiovascular Health and Diseases in China. Report on Cardiovascular Health and Diseases in China 2019: an Updated Summary. Chinese Circulation Journal 2020;35:833-54.
- Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol 2019;20:e262-73. [Crossref] [PubMed]
- Tama BA, Im S, Lee S. Improving an Intelligent Detection System for Coronary Heart Disease Using a Two-Tier Classifier Ensemble. Biomed Res Int 2020;2020:9816142. [Crossref] [PubMed]
- Dinh A, Miertschin S, Young A, et al. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med Inform Decis Mak 2019;19:211. [Crossref] [PubMed]
- Wu Y, Fang Y. Stroke Prediction with Machine Learning Methods among Older Chinese. Int J Environ Res Public Health 2020;17:1828. [Crossref] [PubMed]
- Gooding HC, de Ferranti SD. Cardiovascular risk assessment and cholesterol management in adolescents: getting to the heart of the matter. Curr Opin Pediatr 2010;22:398-404. [Crossref] [PubMed]
- Nivette A, Ribeaud D, Murray A, et al. Non-compliance with COVID-19-related public health measures among young adults in Switzerland: Insights from a longitudinal cohort study. Soc Sci Med 2021;268:113370. [Crossref] [PubMed]
- Liu J, Hong Y, D'Agostino RB Sr, et al. Predictive value for the Chinese population of the Framingham CHD risk assessment tool compared with the Chinese Multi-Provincial Cohort Study. JAMA 2004;291:2591-9. [Crossref] [PubMed]
- Loscalzo J. Harrison's Cardiovascular Medicine, 3rd Edition. 2017.
- Stekhoven DJ, Bühlmann P. MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics 2012;28:112-8. [Crossref] [PubMed]
- Thompson HW, Mera R, Prasad C. The Analysis of Variance (ANOVA). Nutr Neurosci 1999;2:43-55. [Crossref] [PubMed]
- Guyon I, Weston J, Barnhill S, et al. Gene Selection for Cancer Classification Using Support Vector Machines. Mach Learn 2002;46:389-422. [Crossref]
- Kursa M, Rudnicki W. Feature Selection with Boruta Package. J Stat Softw 2010;36:1-13. [Crossref]
- Breiman L. Random Forests. Mach Learn 2001;45:5-32. [Crossref]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2012;12:2825-30.
- Lemaître G, Nogueira F, Aridas C. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Datasets in Machine Learning. J Mach Learn Res 2017;18:1-5.
- Lundberg S, Lee SI. A Unified Approach to Interpreting Model Predictions. NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems 2017;4768-77.
- Wilson PW, D'Agostino RB, Levy D, et al. Prediction of coronary heart disease using risk factor categories. Circulation 1998;97:1837-47. [Crossref] [PubMed]
- Dufouil C, Beiser A, McLure LA, et al. Revised Framingham Stroke Risk Profile to Reflect Temporal Trends. Circulation 2017;135:1145-59. [Crossref] [PubMed]
- Rooke TW, Hirsch AT, Misra S, et al. 2011 ACCF/AHA focused update of the guideline for the management of patients with peripheral artery disease (updating the 2005 guideline): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society for Vascular Medicine, and Society for Vascular Surgery. Catheter Cardiovasc Interv 2012;79:501-31. [Crossref] [PubMed]
- Takashima N, Turin TC, Matsui K, et al. The relationship of brachial-ankle pulse wave velocity to future cardiovascular disease events in the general Japanese population: the Takashima Study. J Hum Hypertens 2014;28:323-7. [Crossref] [PubMed]
- Regitz-Zagrosek V, Kararigas G. Mechanistic Pathways of Sex Differences in Cardiovascular Disease. Physiol Rev 2017;97:1-37. [Crossref] [PubMed]
- Andersson C, Vasan RS. Epidemiology of cardiovascular disease in young individuals. Nat Rev Cardiol 2018;15:230-40. [Crossref] [PubMed]
- Perlstein TS, Creager MA. The ankle-brachial index as a biomarker of cardiovascular risk: it's not just about the legs. Circulation 2009;120:2033-5. [Crossref] [PubMed]
- Mahmood SS, Levy D, Vasan RS, et al. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 2014;383:999-1008. [Crossref] [PubMed]
- Colafella KMM, Denton KM. Sex-specific differences in hypertension and associated cardiovascular disease. Nat Rev Nephrol 2018;14:185-201. [Crossref] [PubMed]
- Sachdeva A, Cannon CP, Deedwania PC, et al. Lipid levels in patients hospitalized with coronary artery disease: an analysis of 136,905 hospitalizations in Get With The Guidelines. Am Heart J 2009;157:111-117.e2. [Crossref] [PubMed]
- Golia E, Limongelli G, Natale F, et al. Inflammation and cardiovascular disease: from pathogenesis to therapeutic target. Curr Atheroscler Rep 2014;16:435. [Crossref] [PubMed]
- Ishizaka N, Ishizaka Y, Toda E, et al. Are serum carcinoembryonic antigen levels associated with carotid atherosclerosis in Japanese men? Arterioscler Thromb Vasc Biol 2008;28:160-5. [Crossref] [PubMed]
- Bae U, Shim JY, Lee HR, et al. Serum carcinoembryonic antigen level is associated with arterial stiffness in healthy Korean adult. Clin Chim Acta 2013;415:286-9. [Crossref] [PubMed]
- Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med 1965;122:467-81. [Crossref] [PubMed]
- Lin JS, Evans CV, Johnson E, et al. Nontraditional Risk Factors in Cardiovascular Disease Risk Assessment: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018;320:281-97. [Crossref] [PubMed]
- Wang Y, Wang JG. Genome-Wide Association Studies of Hypertension and Several Other Cardiovascular Diseases. Pulse (Basel) 2019;6:169-86. [Crossref] [PubMed]
- Witkowski M, Weeks TL, Hazen SL. Gut Microbiota and Cardiovascular Disease. Circ Res 2020;127:553-70. [Crossref] [PubMed]
(English Language Editors: J. Teoh and J. Jones)