
Clinical and resistance characterization of carbapenem-resistant Klebsiella pneumoniae isolated from intensive care units in China
Introduction
Klebsiella pneumoniae (K. pneumoniae), one of the most common gram-negative bacteria of nosocomial infections, has shown generally upward trend in resistance rate to imipenem and meropenem over the past decade, according to data from the China Antimicrobial Surveillance Network (CHINET) (https://www.chinets.com/Data/GermYear). The prevalence of carbapenem-resistant K. pneumoniae (CRKP) isolates increased rapidly, with the national average up to 8.7%, and CRKP represents the growing problem of antibiotic resistance (1). The nasopharyngeal and gastrointestinal colonization rates of K. pneumoniae could be around 19% and 20%, respectively (2). As one of the most common opportunistic pathogens, K. pneumoniae colonization is considered as a starting point for further infection, and for critically ill and immunocompromised patients, especially patients from intensive care unit (ICU), such risk increases. Alarmingly, the detection rate for CRKP patients in ICUs of tertiary hospitals surged from 0% in 2013 to 75% in 2016, which was far above that for non-ICU patients (3). CRKP from the ICU can cause serious respiratory and circulatory infections with high morbidity and mortality, and consequent poorer prognosis (4). Many studies have indicated that the mortality rate of patients with CRKP infection could increase to about 40–50%, and some risk factors, including ICU stays, higher Acute Physiology and Chronic Health Evaluation (APACHE) II scores, or comorbidity with septic shock and congestive heart failure have been identified to be associated with higher mortality of CRKP infection (5,6). Current research on the mechanism of carbapenem resistance has focused on carbapenemase, extended-spectrum beta-lactamases (ESBLs) or Class C carbapenemase (also known as AmpC) genes combined with abnormal expression of outer membrane porin (OMP), efflux pump and biofilm formation (7,8). Understanding the drug resistance mechanism is urgently required for the prevention and treatment of CRKP infection.
Although CRKP has emerged globally, there are regional differences in strains. An observational study in the USA reported geographic variation in the CRKP occurrence rate, which reflected the overall emergence and dissemination (9). In China, the prevalence of CRKP ranges from 0.9% to 23.6% in different provinces (10), and thus drug resistance status could vary endemically (11). Additionally, discrepancies among regions exist in molecular epidemiology. ST258 is the major sequencing type in Europe and the USA, whereas in East Asia, ST11 is dominant. Moreover, the rate of K. pneumoniae carbapenemase (KPC) in carbapenemase-producing strains is lower in Spain than in Greece, and the prevalence of New Delhi metallo-β-lactamase (NDM) in India is particularly high (12). Determining the current resistance situation is essential for guiding rational use of antibiotics, and it is significant to clarify drug resistance mechanism and epidemic trend for nosocomial prevention and control.
In this study, CRKP strains isolated from ICUs in Beijing, Chongqing and Nantong were compared in terms of carbapenem resistance mechanism, sequence types (STs) and genetic relationships. The clinical features of patients infected with CRKP in these three regions were also analyzed. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4323/rc).
Methods
CRKP strain selection
Non-repetitive CRKP strains (at least resistant to one of imipenem, meropenem and ertapenem) from the first tested sample of each ICU patient from January 2019 to December 2021 were collected in three tertiary hospitals in Chongqing, Beijing, and Nantong in the southwest, north, and southeast of China, respectively. All strains were identified by the VITEK2 system; the antimicrobial susceptibility test was conducted by a VITEK2 fully automatic bacteria identification instrument and matching drug sensitivity card. Susceptibility was defined according to the Clinical and Laboratory Standards Institute (CLSI) breakpoints, and Escherichia coli American Type Culture Collection (ATCC) 25922 and Pseudomonas aeruginosa ATCC27853 were used as quality control strains.
Collection of clinical information
The clinical data of patients infected with CRKP were collected by searching online records, including demographic profile (age, sex, length of stay), comorbid conditions [hypertension, diabetes, chronic obstructive pulmonary disease (COPD), malignant tumor, etc.], invasive procedures (tracheotomy, venous catheter, mechanical ventilation, etc.), medications and other relevant information. This clinical research was a retrospective study, and the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University waived the requirement of ethical approval and informed consent, and patients’ information was processed anonymously and confidentially. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Detection of carbapenemase
Class A, B and D types of carbapenemase in the CRKP strains were detected by a modified CarbaNP method (13). Carbapenemases were extracted from bacteria by ultrasonic disruption and high-speed centrifugation. Substrates I–IV all contained red phenol plus ZnSO4, and different type of carbapenemases caused color change in the different substrates [substrate I: no imipenem; substrate II: imipenem; substrate III: imipenem + tazobactam; substrate VI: imipenem + ethylene diamine tetraacetic acid (EDTA); substrate V: imipenem + tazobactam + EDTA]. K. pneumoniae ATCC BAA-1706, ATCC BAA-1705 and ATCC BAA-2146 were the quality control strains.
Identification of resistance genes and OMP genes
DNA templates were extracted by the boiling method. Polymerase chain reaction (PCR) amplification was performed to identify the most common ESBL genes (blaSHV, blaCTX-M, blaTEM), AmpC genes (blaCIT, blaDHA, blaEBC, blaMOX, blaFOX, blaACC), and carbapenemase genes positive for carbapenemase screening, of which the corresponding genes were Class A (blaGES, blaKPC, blaSME, blaIMI, blaBIC), Class B (blaIMP, blaVIM, blaNDM, blaSPM, blaDIM, blaGIM, blaSIM, blaAIM, blaSMB) and Class D (blaOXA-48), using previously reported primers and conditions (14-16). PCR-positive products of resistance genes were sequenced for comparative analysis by NCBI BLAST. OMP genes ompK35 and ompK36 were detected, and the sequence of positive porin amplification products was compared with the wild type CI507 (GenBank: No. FJ577672) and VM522 (GenBank: No. FJ577673) (17).
MLST
The allelic profiles and STs were determined after PCR amplification of the internal fragments of seven K. pneumoniae housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, tonB) and the allele number combination for gene mapping according to BIGSdb-Pasteur database (https://bigsdb.pasteur.fr/). Primer design and the PCR process were referred to the MLST website (https://bigsdb.pasteur.fr/klebsiella/primers_used.html), and genetic relationships were analyzed by PHYLOViZ software.
Statistical analysis
The regional comparison was performed by IBM SPSS26 (USA). Chi-square test were used to determine the differences in drug resistance rate, resistance genes and major STs, and P values were calculated by Pearson chi-square test, continuity correction or Fisher’s exact test. Clinical profiles included in this study were analyzed retrospectively. All variables were described as qualitative data. Two-sided P values <0.05 were considered statistically significant, and the Bonferroni-corrected P value for pairwise comparisons was 0.05/3=0.0167.
Results
Comparison of clinical characteristics
The demographic and clinical profiles of patients infected with CRKP are shown in Table 1. Of these 135 patients (complete data of 15 patients was unavailable), 71.85% were ≥60 years, and the number of male patients was almost 3-fold more than that of female patients. Nearly half of the patients had been in the ICU for ≥7 days, and had been hospitalized within 2 months before ICU admission. The highest incidence of underlying disease was hypertension and chronic cardiac insufficiency in Chongqing and Beijing, respectively, and hypertension and diabetes in Nantong. Most of the patients had received ventilatory assistance, central venous catheterization, sputum suction and other operations. Carbapenems were the most frequently used antibiotics in all three hospitals, and more than 50% of the patients were co-infected with other bacteria, at higher rates in Chongqing and Beijing.
Table 1
| Characteristics | Total (n=135) | Chongqing (n=52) | Nantong (n=36) | Beijing (n=47) | P valuea | P valueb | P valuec | P value |
|---|---|---|---|---|---|---|---|---|
| Sex (male) | 100 (74.07) | 38 (73.08) | 28 (77.78) | 34 (72.34) | 0.617 | 0.935 | 0.572 | 0.836 |
| Age (≥60 years) | 97 (71.85) | 31 (59.62) | 27 (75.00) | 39 (82.98) | 0.134 | 0.011* | 0.372 | 0.032* |
| Hospital stay (in 2 months) | 78 (57.78) | 26 (50.00) | 21 (58.33) | 31 (65.96) | 0.441 | 0.109 | 0.477 | 0.275 |
| Length of ICU stay before CRKP isolation (≥7 days) | 72 (53.33) | 26 (50.00) | 18 (50.00) | 28 (59.57) | 1.000 | 0.339 | 0.384 | 0.569 |
| Comorbidities | ||||||||
| Hypertension | 47 (34.81) | 19 (36.54) | 16 (44.44) | 12 (25.53) | 0.456 | 0.238 | 0.071 | 0.19 |
| Diabetes mellitus | 44 (32.59) | 8 (15.38) | 16 (44.44) | 20 (42.55) | 0.003* | 0.003* | 0.863 | 0.003* |
| Chronic heart failure | 62 (45.93) | 19 (36.54) | 7 (19.44) | 36 (76.60) | 0.084 | <0.001* | <0.001* | <0.001* |
| COPD | 14 (10.37) | 4 (7.69) | 3 (8.33) | 7 (14.89) | 1.000 | 0.255 | 0.569 | 0.459 |
| Chronic renal disease | 26 (19.26) | 12 (23.08) | 5 (13.89) | 9 (19.15) | 0.283 | 0.633 | 0.526 | 0.561 |
| Chronic hepatopathy | 29 (21.48) | 18 (34.62) | 3 (8.33) | 8 (17.02) | 0.004* | 0.047 | 0.406 | 0.008* |
| Malignant tumor | 22 (16.30) | 5 (9.62) | 4 (11.11) | 13 (27.66) | 1.000 | 0.02 | 0.064 | 0.043* |
| Impaired immune system | 12 (8.89) | 6 (11.54) | 1 (2.78) | 5 (10.64) | 0.233 | 0.887 | 0.227 | 0.349 |
| Neutropenia | 10 (7.41) | 5 (9.62) | 1 (2.78) | 4 (8.51) | 0.394 | 1.000 | 0.382 | 0.502 |
| Invasive operations | ||||||||
| Tracheotomy | 46 (34.07) | 12 (23.08) | 24 (66.67) | 10 (21.28) | <0.001* | 0.83 | <0.001* | <0.001* |
| Mechanical ventilation | 102 (75.56) | 42 (80.77) | 24 (66.67) | 36 (76.60) | 0.133 | 0.612 | 0.317 | 0.311 |
| Central venous catheter | 117 (86.67) | 46 (88.46) | 29 (80.56) | 42 (89.36) | 0.304 | 0.887 | 0.258 | 0.448 |
| Urinary catheter | 122 (90.37) | 46 (88.46) | 33 (91.67) | 43 (91.49) | 0.896 | 0.869 | 1.000 | 0.873 |
| Sputum suction | 102 (75.56) | 38 (73.08) | 24 (66.67) | 40 (85.11) | 0.517 | 0.144 | 0.048 | 0.133 |
| Enteral nutrition | 97 (71.85) | 34 (65.38) | 30 (83.33) | 33 (70.21) | 0.063 | 0.608 | 0.166 | 0.841 |
| Parenteral nutrition | – | 10 (19.23) | 4 (11.11) | – | 0.306 | – | – | – |
| Immunosuppressant | – | 2 (3.85) | 0 (0.00) | – | 0.511 | – | – | – |
| Hormonotherapy | – | 34 (65.38) | – | – | – | – | – | – |
| Antibiotics | ||||||||
| Cephalosporins | 45 (33.33) | 20 (38.46) | 11 (30.56) | 14 (29.79) | 0.445 | 0.364 | 0.94 | 0.292 |
| Carbapenems | 69 (51.11) | 34 (65.38) | 20 (55.56) | 15 (31.91) | 0.352 | 0.001* | 0.031 | 0.003* |
| Aminoglycosides | 7 (5.19) | 3 (5.77) | 1 (2.78) | 3 (6.38) | 0.642 | 1.000 | 0.629 | 0.793 |
| Quinolones | 29 (21.48) | 13 (25.00) | 3 (8.33) | 13 (27.66) | 0.046 | 0.764 | 0.027 | 0.077 |
| Penicillins | 48 (35.56) | 20 (38.46) | 17 (47.22) | 11 (23.40) | 0.413 | 0.107 | 0.023 | 0.064 |
| Glycopeptides | 31 (22.96) | 17 (32.69) | 3 (8.33) | 11 (23.40) | 0.007* | 0.306 | 0.069 | 0.028* |
| Linezolid | 32 (23.70) | 19 (36.54) | 0 (0.00) | 13 (27.66) | <0.001* | 0.346 | 0.001* | <0.001* |
| Tigecycline | 28 (20.74) | 11 (21.15) | 7 (19.44) | 10 (21.28) | 0.845 | 0.988 | 0.838 | 0.975 |
| Antifungal agents | 44 (32.59) | 29 (55.77) | 9 (25.00) | 6 (12.77) | 0.004* | <0.001* | 0.151 | <0.001* |
| Infection | ||||||||
| Fungal infection | 21 (15.56) | 12 (23.08) | 6 (16.67) | 3 (6.38) | 0.464 | 0.021 | 0.255 | 0.056 |
| Other bacterial infections | 108 (80.00) | 46 (88.46) | 20 (55.56) | 42 (89.36) | <0.001* | 0.887 | <0.001* | <0.001* |
Data are present as n (%). P valuea: Chongqing vs. Nantong; P valueb: Chongqing vs. Beijing; P valuec: Nantong vs. Beijing; P value: Chongqing vs. Nantong vs. Beijing; –: not available. *P<0.05. ICU, intensive care unit; CRKP, carbapenem-resistant Klebsiella pneumoniae; COPD, chronic obstructive pulmonary disease.
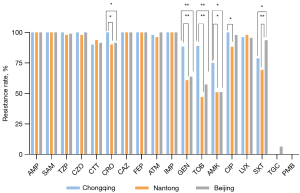
Antimicrobial susceptibility testing of CRKP strains
Totals of 52 strains from Chongqing, 51 strains from Nantong and 47 strains from Beijing were collected, and clinical samples were sputum (n=106), urine (n=22), ascites (n=10), pus (n=4) and other (n=8). In addition to 100% resistance to imipenem, CRKP strains were extensively resistant to other antibiotics (Figure 1). The resistance rates of strains to penicillins (ampicillin, ampicillin sulbactam, piperacillin tazobactam), cephalosporins (cefazolin, cefotetan, ceftazidime, cefepime), aztreonam and levofloxacin were extremely high (≥90%) in all ICUs. Different to Chongqing, the resistance rate of aminoglycosides (gentamicin, tobramycin, amikacin) was much lower in Nantong and Beijing. Moreover, tigecycline-resistant strains were only detected in Beijing and all strains were susceptible to polymyxin B.
Resistance mechanism of carbapenems
In this study, 100 strains producing Class A carbapenemases carried blaKPC-2 and 8 strains producing Class B carbapenemases carried blaNDM-1 (Figure 2). There were no strains producing both A and B, or D carbapenemases. For CRKP from Chongqing, 35 strains carried blaKPC-2 and 4 strains carried blaNDM-1; for strains from Nantong, 28 carried blaKPC-2 and 1 carried blaNDM-1; and for strains from Beijing, 28 carried blaKPC-2 and 1 carried blaNDM-1. Other related carbapenemase genes were not found in our study. Both AmpC and ESBL genes were identified, of which the prevalence rates were higher in Nantong. A total of 3 strains with ompK35 deletion were found from Nantong, and ompK36 deletion was detected, comprising 2 from Chongqing and 2 from Nantong. Notably, 1 strain simultaneously producing KPC carbapenemase, AmpCs and ESBLs did not express the ompK36 gene, and another strain producing NDM carbapenemase and ESBLs was missing OmpK35. Both these strains were highly resistant to imipenem. Resistance mechanisms showed few differences except for expression of AmpCs and ESBLs genes (Table 2).
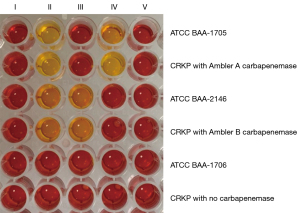
Table 2
| Resistance mechanism | Chongqing (n=52) | Nantong (n=51) | Beijing (n=47) | P valuea | P valueb | P valuec | P value |
|---|---|---|---|---|---|---|---|
| Resistance genes | |||||||
| blaKPC-2 | 35 (67.31) | 28 (54.9) | 37 (78.72) | 0.196 | 0.203 | 0.013* | 0.044* |
| blaNDM-1 | 4 (7.69) | 1 (1.96) | 3 (6.38) | 0.363 | 1.000 | 0.347 | 0.437 |
| blaDHA | 2 (3.85) | 16 (31.37) | 8 (17.02) | <0.001* | 0.066 | 0.099 | 0.001* |
| blaCIT | 1 (1.92) | 0 (0.00) | 0 (0.00) | 1.000 | 1.000 | – | 1.000 |
| blaSHV | 48 (92.31) | 51 (100.00) | 33 (70.21) | 0.118 | 0.004* | <0.001* | <0.001* |
| blaTEM | 50 (96.15) | 51 (100.00) | 37 (78.72) | 0.495 | 0.008* | 0.001* | <0.001* |
| blaCTX-M | 38 (73.08) | 50 (98.04) | 31 (65.96) | <0.001* | 0.441 | <0.001* | <0.001* |
| OMP loss | 2 (3.85) | 5 (9.80) | 0 (0.00) | 0.269 | 0.496 | 0.057 | 0.06 |
| Carbapenemases+ | |||||||
| Class A, AmpCs+, ESBLs+, OMP loss | 1 (1.92) | 0 (0.00) | 0 (0.00) | 1.000 | 1.000 | – | 1.000 |
| Class B, ESBLs+, OMP loss | 0 (0.00) | 1 (1.96) | 0 (0.00) | 0.495 | – | 1.000 | 0.653 |
| Class A, AmpCs+, ESBLs+ | 2 (3.85) | 9 (17.65) | 1 (2.13) | 0.023 | 1.000 | 0.028 | 0.013* |
| Class A, AmpCs+, ESBLs− | 0 (0.00) | 0 (0.00) | 1 (2.13) | – | 0.475 | 0.48 | 0.313 |
| Class A, AmpCs−, ESBLs+ | 33 (63.46) | 19 (37.25) | 32 (68.09) | 0.008* | 0.629 | 0.002* | 0.004* |
| Class A, AmpCs−, ESBLs− | 0 (0.00) | 0 (0.00) | 3 (6.38) | – | 0.103 | 0.107 | 0.029* |
| Class B, AmpCs+, ESBLs+ | 0 (0.00) | 0 (0.00) | 0 (0.00) | – | – | – | – |
| Class B, AmpCs+, ESBLs− | 0 (0.00) | 0 (0.00) | 0 (0.00) | – | – | – | – |
| Class B, AmpCs−, ESBLs+ | 4 (7.69) | 1 (1.96) | 3 (6.38) | 0.363 | 1.000 | 0.347 | 0.437 |
| Class B, AmpCs−, ESBLs− | 0 (0.00) | 0 (0.00) | 0 (0.00) | – | – | – | – |
| Carbapenemases− | |||||||
| AmpCs+ | 1 (1.92) | 7 (13.73) | 7 (14.89) | 0.031 | 0.025 | 0.869 | 0.038* |
| ESBLs+ | 13 (25.00) | 22 (43.14) | 10 (21.28) | 0.052 | 0.661 | 0.021 | 0.039* |
| AmpCs+, OMP loss | 0 (0.00) | 1 (1.96) | 0 (0.00) | 0.495 | – | 1.000 | 0.653 |
| ESBLs+, OMP loss | 0 (0.00) | 1 (1.96) | 0 (0.00) | 0.495 | – | 1.000 | 0.653 |
| AmpCs+, ESBLs+ | 1 (1.92) | 7 (13.73) | 7 (14.89) | 0.031 | 0.025 | 0.869 | 0.055 |
| AmpCs+, ESBLs+, OMP loss | 0 (0.00) | 1 (1.96) | 0 (0.00) | 0.495 | – | 1.000 | 0.653 |
Data are present as n (%). P valuea: Chongqing vs. Nantong; P valueb: Chongqing vs. Beijing; P valuec: Nantong vs. Beijing; P value: Chongqing vs. Nantong vs. Beijing. –: not available. *P<0.05. OMP, outer membrane porin; ESBLs, extended-spectrum beta-lactamase genes; AmpCs, Class C carbapenemase genes.
Molecular epidemiology
In this study, there were altogether 28 STs among the 150 CRKP strains from the three regions, mainly ST11 (76/150, 50.67%), ST15 (33/150, 22%), ST13 (6/150, 4%), ST1 (3/150, 2%), ST48 (3/150, 2%), and several other types (Figure 3). Among 52 CRKP strains from Chongqing, ST11 was major (31/52, 59.6%), followed by ST15 (9/52, 17.3%). For the 51 strains from Nantong, ST11 (21/51, 41.8%) and ST15 (21/51, 41.8%) were the most common. And of the 47 strains from Beijing, ST11 was dominant (24/47, 51.06%). There was no difference among the three regions in the composition ratio of ST11, but the difference of ST15 was statistically significant between Nantong and both Chongqing and Beijing (P=0.008 & P<0.001). BURST analysis showed that ST11, ST340, ST859, ST1326, ST1953, ST690 and ST1883 belonged to clonal complex 11, whereas ST37 and ST2020, ST1561 and ST147, ST48 and ST2361 belonged to three different sequence groups respectively, and the other types were singletons (Figure 4).


Discussion
Admission to an ICU is one of the risk factors for CRKP infection (18), and the morbidity of CRKP infection in ICU patients is higher than for other patients (19). A 4-year report revealed that, without paying attention to hand hygiene and environmental cleaning and disinfection, it was ineffective to isolate patients infected with CRKP. The hand hygiene and environmental cleaning were however, important, because CRKP could be cross-transmitted through the hands of medical staff and patients’ surroundings (20). Moreover, for patients with coronavirus disease 2019 (COVID-19) patients, the prevalence of CRKP ranged from 0.35–53%, and because of mechanical ventilation and central catheters in ICUs, pulmonary and bloodstream infection were the main types (21). In this study, the characteristics of patients infected with CRKP were roughly similar to those in previous studies (22,23). There was no significant difference in most of the patients in the three study hospitals, apart from the underlying diseases and use of antibiotics, which were mainly due to the hospital admission and antibiotics available. Other than K. pneumoniae, the most common bacteria detected in this study were P. aeruginosa and Acinetobacter baumannii, and the isolation rates of the latter two species were higher in Chongqing and Beijing. Despite the advanced medical equipment and better infection control in tertiary hospitals, the patients admitted to the ICU of a tertiary hospitals generally suffer more serious disease or have been transferred from secondary hospitals, thus the risk of co-infection increased (24). In their analysis of the risk factors for colonization of ICU pathogens, a Greek study reported that KPC-producing K. pneumoniae and vancomycin-resistant Enterococcus were important risk factors for each other’s colonization, which proved the risk of CRKP infection for the colonization of other multidrug-resistant pathogens in that institute (25). In general, patients from three ICUs were critically ill and treated with multiple antibiotics, increasing the risk of CRKP infection. At the same time, it also increased the risk of infection by other drug-resistant bacteria and fungal infection.
The CRKP strains in this study were highly resistant to most antibiotics, which may be related to the duration and various types of antibiotic exposure, and widespread of antibiotic-resistance genes (26). Moreover, resistance rates basically showed a slight increase from previous local reports (27,28). It is worth noting that the resistance rate of CRKP to aminoglycosides was comparatively lower, consistent with the current status of drug resistance (29). The low resistance rate of aminoglycosides may be due to their adverse reactions and thus lower use in clinical practice, suggesting that scheduled shifting or switching of empirical medication might alleviate antibiotic resistance (30). Furthermore, tigecycline and polymyxin B are still the most effective drugs against CRKP infection (31). A previous report showed that the resistance rates of K. pneumoniae in the Jiangzhe area were higher than in the southwest regions of China, which was contrary to this study’s findings (32). As to the drug resistance in the three study hospitals, there were different antibiotics available and used, especially simultaneously, the locality of positioning of the hospitals was different, and the admitted patients were also different in their characteristics. Under these many different external conditions, there was no significant difference in the separated CRKP stains in the three hospitals.
Production of carbapenemases is the most important mechanism of carbapenem resistance of K. pneumoniae. Only blaKPC-2 and blaNDM-1 were found in this study. The main carbapenemase gene in Chongqing and Nantong was blaKPC-2, which was similar to previous studies (33,34). blaKPC-2 was also the major carbapenemase gene in Beijing, although previous research showed the reverse with predominance of blaNDM-1 in Beijing (35), for which the reason might be that wide dissemination of KPC-producing K. pneumoniae has already happened in China (12). Although the detection rate of blaNDM was relatively low in China, it is predominant in India and Pakistan (36,37). Domestic travel or transportation of patients infected with carbapenemase-producing K. pneumoniae might be the primary reason for its dissemination (12). A study in the United Kingdom verified that the epidemic of CRKP in Europe was driven by several carbapenemase-producing clonal lineages, so rapid dissemination could be also attributed to mobile resistance elements, and thus closer domestic and international communication increases the risk of CRKP transmission (38,39). Additionally, ESBL genes were prevalent in the CRKP strains and nearly all strains in this study harbored one or two ESBL genes (blaSHV, blaTEM, blaCTX-M), and the prevalence rate was close to related reports in Chongqing, Jiangsu, and Beijing (35-37). As an important part of the outer membrane of bacteria, porin plays a key role in maintaining membrane permeability and regulating the entry and exit of substances. In this study, OMP genes (ompK35/ompK36) of CRKP were directly examined by PCR, and loss of porin existed in a small number of strains only in Chongqing and Nantong. Additionally, point and frameshift mutations were found in both ompK35 and ompK36 genes, which might result in abnormal expression of OMP, thus leading to drug resistance. Research has indicated that some strains producing no carbapenenase but lacking porin with ESBL- or AmpC-positive also showed resistance to imipenem, which seems to confirm the substitution mechanism of carbapenem resistance (40,41). Moreover, other mechanisms such as overexpression of the efflux pump and biofilm formation could also contribute to carbapenem resistance, which explains the result of antibiotic susceptibility that strains without resistance genes were also resistant to imipenem. Generally speaking, K. pneumoniae from the three ICUs developed resistance to carbapenem mainly by producing carbapenemases and other enzymes, which means more careful and appropriate antibiotic use should be considered. According to our experimental results the drug resistance mechanism of CRKP in the three hospitals tended to be similar.
The polyclonal spread of CRKP can be identified by MLST or pulsed field gel electrophoresis (PFGE), analyzing the major lineages of isolates could suggest whether local clonal expansions exist. ST11 (50.67%) and ST15 (22%) were the main genotypes in this study. ST11 is predominant in China and East Asia. ST15 differs from ST11 by only two alleles. ST11 K. pneumoniae possesses pathogenic potential and resistance to serum killing, so an outbreak would be difficult to control (42). The main STs in the three hospitals were slightly different, ST11 and ST15 were dominant in Nantong, while in Chongqing and Beijing only ST11 was the leading genotype, which proved the regional differentiation of major STs. There are few reports on ST15 K. pneumoniae in Nantong, however, a study revealed that ST15 was common in Shanghai, a city geographically close to Nantong, and the major STs might be related to interregional transmission (43). Among the 150 CRKP, blaKPC-2 carriers were mainly ST11. Moreover, some other STs also harbored blaKPC or blaNDM, which proved horizontal transmission of carbapenemase genes (44). Other less common STs were found in this study. Taken together, despite the discrepancy in rarer genotypes, the major types of the three hospitals were largely consistent.
Despite the three cities being geographically far apart, and the differing clinical characteristics of the patients with CRKP infection, the resistance profiles and resistance mechanism of CRKP essentially coincided with few differences. The blaKPC resistance gene has faster and wider transmission. In order to better control the clinical prevalence of CRKP, its transmission mechanism is worthy of in-depth study.
In conclusion, our study indicated that under the premise of sufficient movement of people across the country, the dominant resistant strains of K. pneumoniae are widely distributed, which causes the prevalence of resistance strains in different cities to be consistent. Although the resistance mechanism of CRKP was not investigated thoroughly in this study and the sample size was small, it has provided some theoretical evidence for infection control and support for considered application of antibiotics.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Chongqing (grant No. cstc2020jcyj-msxmX0415) and Kuanren Talents Program of The Second Affiliated Hospital of Chongqing Medical University.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4323/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4323/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4323/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013). This clinical study was a retrospective study, and the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University waived the requirement of ethical approval and informed consent, and patients’ information was processed anonymously and confidentially.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hu F, Zhu D, Wang F, et al. Current Status and Trends of Antibacterial Resistance in China. Clin Infect Dis 2018;67:S128-34. [Crossref] [PubMed]
- Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018;8:4. [Crossref] [PubMed]
- Tian L, Zhang Z, Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: a 20-year surveillance study (1998-2017). Antimicrob Resist Infect Control 2019;8:86. [Crossref] [PubMed]
- Wang Z, Qin RR, Huang L, et al. Risk Factors for Carbapenem-resistant Klebsiella pneumoniae Infection and Mortality of Klebsiella pneumoniae Infection. Chin Med J (Engl) 2018;131:56-62. [Crossref] [PubMed]
- Wu C, Zheng L, Yao J. Analysis of Risk Factors and Mortality of Patients with Carbapenem-Resistant Klebsiella pneumoniae Infection. Infect Drug Resist 2022;15:2383-91. [Crossref] [PubMed]
- Qian Y, Bi Y, Liu S, et al. Predictors of mortality in patients with carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis and a systematic review. Ann Palliat Med 2021;10:7340-50. [Crossref] [PubMed]
- Tooke CL, Hinchliffe P, Bragginton EC, et al. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J Mol Biol 2019;431:3472-500. [Crossref] [PubMed]
- Lutgring JD, Limbago BM. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J Clin Microbiol 2016;54:529-34. [Crossref] [PubMed]
- Han JH, Goldstein EJ, Wise J, et al. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Network of Long-Term Acute Care Hospitals. Clin Infect Dis 2017;64:839-44. [PubMed]
- Zhou M, Wang Y, Liu C, et al. Comparison of five commonly used automated susceptibility testing methods for accuracy in the China Antimicrobial Resistance Surveillance System (CARSS) hospitals. Infect Drug Resist 2018;11:1347-58. [Crossref] [PubMed]
- Wang N, Zhan M, Liu J, et al. Prevalence of Carbapenem-Resistant Klebsiella pneumoniae Infection in a Northern Province in China: Clinical Characteristics, Drug Resistance, and Geographic Distribution. Infect Drug Resist 2022;15:569-79. [Crossref] [PubMed]
- Lee CR, Lee JH, Park KS, et al. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol 2016;7:895. [Crossref] [PubMed]
- Dortet L, Poirel L, Nordmann P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 2012;56:6437-40. [Crossref] [PubMed]
- Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 2002;40:2153-62. [Crossref] [PubMed]
- Dallenne C, Da Costa A, Decré D, et al. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010;65:490-5. [Crossref] [PubMed]
- Kaczmarek FM, Dib-Hajj F, Shang W, et al. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother 2006;50:3396-406. [Crossref] [PubMed]
- Song W, Suh B, Choi JY, et al. In vivo selection of carbapenem-resistant Klebsiella pneumoniae by OmpK36 loss during meropenem treatment. Diagn Microbiol Infect Dis 2009;65:447-9. [Crossref] [PubMed]
- Tian L, Tan R, Chen Y, et al. Epidemiology of Klebsiella pneumoniae bloodstream infections in a teaching hospital: factors related to the carbapenem resistance and patient mortality. Antimicrob Resist Infect Control 2016;5:48. [Crossref] [PubMed]
- Jiang ZQ, Wang SD, Feng DD, et al. Epidemiological risk factors for nosocomial bloodstream infections: A four-year retrospective study in China. J Crit Care 2019;52:92-6. [Crossref] [PubMed]
- Cohen MJ, Block C, Levin PD, et al. Institutional control measures to curtail the epidemic spread of carbapenem-resistant Klebsiella pneumoniae: a 4-year perspective. Infect Control Hosp Epidemiol 2011;32:673-8. [Crossref] [PubMed]
- Mędrzycka-Dąbrowska W, Lange S, Zorena K, et al. Carbapenem-Resistant Klebsiella pneumoniae Infections in ICU COVID-19 Patients-A Scoping Review. J Clin Med 2021;10:2067. [Crossref] [PubMed]
- Colodner R, Rock W, Chazan B, et al. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis 2004;23:163-7. [Crossref] [PubMed]
- Liu P, Li X, Luo M, et al. Risk Factors for Carbapenem-Resistant Klebsiella pneumoniae Infection: A Meta-Analysis. Microb Drug Resist 2018;24:190-8. [Crossref] [PubMed]
- Fatovich DM, Phillips M, Jacobs IG. A comparison of major trauma patients transported to trauma centres vs. non-trauma centres in metropolitan Perth. Resuscitation 2011;82:560-3. [Crossref] [PubMed]
- Papadimitriou-Olivgeris M, Spiliopoulou I, Christofidou M, et al. Co-colonization by multidrug-resistant bacteria in two Greek intensive care units. Eur J Clin Microbiol Infect Dis 2015;34:1947-55. [Crossref] [PubMed]
- Gorrie CL, Mirceta M, Wick RR, et al. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin Infect Dis 2017;65:208-15. [Crossref] [PubMed]
- Gong YL, Liu CJ, Luo XQ, et al. Epidemiology investigation of carbapenems-resistant Klebsiella pneumoniae in burn care unit. Zhonghua Shao Shang Za Zhi 2019;35:798-803. [PubMed]
- Chen Z, Xiang J. Preliminary study on resistance mechanism and virulence features in carbapenems-resistant Klebsiella pneumoniae from burn patients. Zhonghua Shao Shang Za Zhi 2018;34:796-801. [PubMed]
- Huang W, Zhang J, Zeng L, et al. Carbapenemase Production and Epidemiological Characteristics of Carbapenem-Resistant Klebsiella pneumoniae in Western Chongqing, China. Front Cell Infect Microbiol 2021;11:775740. [Crossref] [PubMed]
- Chatzopoulou M, Reynolds L. Systematic review of the effects of antimicrobial cycling on bacterial resistance rates within hospital settings. Br J Clin Pharmacol 2022;88:897-910. [Crossref] [PubMed]
- Tian Y, Zhang Q, Wen L, et al. Combined effect of Polymyxin B and Tigecycline to overcome Heteroresistance in Carbapenem-Resistant Klebsiella pneumoniae. Microbiol Spectr 2021;9:e0015221. [Crossref] [PubMed]
- Zhang H, Zhang G, Yang Y, et al. Antimicrobial resistance comparison of Klebsiella pneumoniae pathogens isolated from intra-abdominal and urinary tract infections in different organs, hospital departments and regions of China between 2014 and 2017. J Microbiol Immunol Infect 2021;54:639-48. [Crossref] [PubMed]
- Lin Q, Wu M, Yu H, et al. Clinical and Microbiological Characterization of Carbapenem-Resistant Enterobacteriales: A Prospective Cohort Study. Front Pharmacol 2021;12:716324. [Crossref] [PubMed]
- Zhu WM, Yuan Z, Zhou HY. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection relative to two types of control patients: a systematic review and meta-analysis. Antimicrob Resist Infect Control 2020;9:23. [Crossref] [PubMed]
- Dong F, Zhang Y, Yao K, et al. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae Bloodstream Infections in a Chinese Children's Hospital: Predominance of New Delhi Metallo-β-Lactamase-1. Microb Drug Resist 2018;24:154-60. [Crossref] [PubMed]
- Yan J, Pu S, Jia X, et al. Multidrug Resistance Mechanisms of Carbapenem Resistant Klebsiella pneumoniae Strains Isolated in Chongqing, China. Ann Lab Med 2017;37:398-407. [Crossref] [PubMed]
- Gu B, Bi R, Cao X, et al. Clonal dissemination of KPC-2-producing Klebsiella pneumoniae ST11 and ST48 clone among multiple departments in a tertiary teaching hospital in Jiangsu Province, China. Ann Transl Med 2019;7:716. [Crossref] [PubMed]
- Martin J, Phan HTT, Findlay J, et al. Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission. J Antimicrob Chemother 2017;72:3025-34. [Crossref] [PubMed]
- David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 2019;4:1919-29. [Crossref] [PubMed]
- van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020;20:731-41. [Crossref] [PubMed]
- Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, et al. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents 2018;52:898-905. [Crossref] [PubMed]
- Wei DD, Wan LG, Deng Q, et al. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis 2016;85:192-4. [Crossref] [PubMed]
- Li X, Ma W, Qin Q, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol 2019;19:235. [Crossref] [PubMed]
- Giakkoupi P, Papagiannitsis CC, Miriagou V, et al. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J Antimicrob Chemother 2011;66:1510-3. [Crossref] [PubMed]
(English Language Editor: K. Brown)


