
Hedysarum multijugum Maxim treats ulcerative colitis through the PI3K-AKT and TNF signaling pathway according to network pharmacology and molecular docking
Introduction
Ulcerative colitis (UC) is a chronic inflammatory bowel disease (IBD) that impairs the colon and rectal mucosae; to date, no recognized cause has been confirmed (1). Based on recent studies, patients with UC probably have genetic and environmental predispositions, intestinal microenvironment dysregulation, and immunity disorder. An inability to distinguish pathogens and commensal microbes, overactivated innate and adaptive immune responses, and weak inhibitory immune regulation are the potential pathophysiology of UC (2,3). Pathological changes are limited to the mucosal membrane and submucosal layer of the colon and rectum, which are distributed continuously and diffusely. Bloody diarrhea is the major symptom of UC, and abdominal pain is experienced by UC patients with mild symptoms. The aim of treatment is to maintain symptom relief and mucosal healing, prevent complications, and improve the quality of life of patients. Mesalazine, corticosteroids, and immunosuppressive drugs are usually applied to control the inflammatory response. Symptomatic treatment is used to maintain the patient’s water and electrolyte balance. Surgery is required for patients with complications or severe symptoms. The currently available treatments either have limited effects or safety problems (4).
Traditional Chinese medicine (TCM) treatment has been shown to have incredible effects on UC (5-7). Hedysarum multijugum Maxim (HMM), a type of traditional Chinese herb which is mainly distributed in central China, has long been used to treat UC. In general, HMM is used for ulceration and to improve immunity. Previous research has revealed that HMM has good effects on treating UC (8,9). However, the mechanism of HMM treating UC needs further exploration. Therefore, in the present research, network pharmacology and molecular docking were applied to explore the effect target of UC and active ingredients of HMM, not only explaining the underlying mechanism but providing a direction for drug innovation in the future.
The HMM targeting genes were collected according to HMM’s active ingredients and intersected with UC-related genes. Enriched functions and signaling pathways were explored by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Besides, a network comprising significant genes, active compounds, and corresponding signaling pathways was visualized by software such as Cytoscape (https://cytoscape.org/) or R package (https://www.r-project.org/). This technology helps researchers to study the effect of multiple compounds on different genes at one time, providing a comprehensive understanding of the mechanism of disease treatment (10). Additionally, molecular docking was first used to discover molecular interactions and now is applied to drug discovery (11). In this study, molecular docking was executed to predict the binding region of ingredients and receptors and to assess their binding affinity. In recent years, there has been plenty of network pharmacology research conducted focusing on the treatment of chronic diseases with TCM (12). According to the findings of this study, HMM treats UC by modulating immune response and inflammation via the PI3K-AKT and tumor necrosis factor (TNF) signaling pathways. This research might establish a solid foundation for future research and therapeutic innovation. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4815/rc).
Methods
Acquisition of the active ingredients and target genes of HMM
Ingredients of HMM were obtained from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (13) (TCMSP; http://tcmspw.com/). The parameters of oral bioavailability (OB) greater than 30%, and drug-likeness (DL) greater than 0.18 were applied to screen active ingredients (14). The OB reflects the proportion of the amount of drug absorbed by blood circulation (15). The DL refers to the similarity between ingredients and known drugs. Target genes related to active ingredients were selected from the TCMSP database (13). Target gene symbols were annotated by gene information from Uniport (16) (http://www.uniprot.org/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Obtainment of a UC-related gene set
Three databases were used to search genes related to UC: Genecards database (17) (https://www.genecards.org/), PharmGkb database (18) (https://www.pharmgkb.org/), and Therapeutic Target Database (TTD) database (19) (http://db.idrblab.net/ttd/). Genes acquired from the Genecards database were filtered by relevance score. Genes with relevance scores greater than 1 were retained. The common genes of these three databases comprised the UC-related gene set.
Analysis of overlapping genes between HMM and UC
Candidate genes screening
The candidate genes were the intersection of HMM target genes and UC-related genes. They were screened through R software (version 4.0.4) and a Venn diagram was drawn using the VennDiagram package in R.
Protein-protein interaction (PPI) network and compound-candidate gene network
PPI networks were created by the Search Tool for the Retrieval of Interacting Genes/genomes (STRING); (https://string-db.org) database based on candidate genes data, with the highest confidence selected and disconnected nodes hidden (20). A bar plot exhibiting the most frequently interacted proteins among candidate genes was created by R software (version 4.0.4). Cytoscape 3.8.2, a platform for integrating biomolecular interaction networks, was used to construct a compound candidate genes network (21).
GO and KEGG enrichment analysis
We performed GO and KEGG enrichment analyses to evaluate the biological processes (BP), cellular components (CC), molecular functions (MF), and signaling pathways that candidate genes potentially regulated. For these two enrichment analyses, the thresholds of the P value and q-value were both 0.05. Enrichment analyses were carried out by the Cluster profile package in R version 4.0.4 (22).
Hub genes screening and compound-hub genes-signaling pathway construction
Betweenness, Closeness, Degree, Eigenvector, LAC, and Network of each gene were calculated via the CytoNca plugin in Cytoscape (23,24). The primary subnetwork comprised candidate genes with all parameters greater than the median value. Hub genes were selected from genes screened from the primary subnetwork under the same screening parameters. The top 10 signaling pathways, hub genes, and corresponding compounds were connected, and the averages of the respective degrees were set as the threshold values to explore the significant compounds, candidate genes, and signaling pathways that comprised the critical network.
Molecular docking simulation
Ligand preparation
Two-dimensional (2D) structures of active ingredients were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and were transformed into three-dimensional (3D) structures by ChemBio 3D (25). The energy of the 3D structure was minimized for optimization.
Target protein preparation
The 3D structure of receptor proteins encoded by candidate genes was downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (http://www.pdb.org/) (26). Water molecules and organic ligands of receptor proteins were removed by PyMol2.4.2. Hydrogenation and charge calculation on receptor proteins were performed by Autodock Tools (27). Active pocket sites of receptor proteins were searched based on settings.
Molecular docking
Based on active pocket sites, molecular docking of compound-target was performed by Autodock Vina. Compound-target binding models were then visualized by PyMol2.4.2.
Statistical analysis
All statistical analyses were conducted with R software (Version 3.6.1). A P value of less than 0.05 was regarded as statistically significant unless otherwise stated, and all P values were two tailed.
Results
Active ingredients and target genes screening
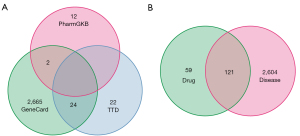
The flowchart of this study is exhibited in Figure 1. 20 active ingredients and 180 target genes of HMM were obtained from the TCMSP database (Table S1 and https://cdn.amegroups.cn/static/public/atm-22-4815-1.xlsx). Then, 2,691, 46, and 14 genes related to UC were obtained from the Genecards database, TTD database, and PharmGKB database, respectively (Figure 2A). There were 2,725 UC-related genes after screening (https://cdn.amegroups.cn/static/public/atm-22-4815-2.xlsx). The intersecting parts of compound target genes and UC-related genes were taken (https://cdn.amegroups.cn/static/public/atm-22-4815-3.xlsx), and in the end, 121 candidate genes were preserved (Figure 2B).
PPI and candidate genes-compound network
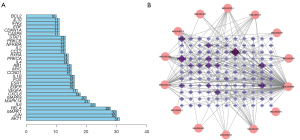
Processed by the STRING database, the PPI network showed complicated interactions between candidate genes at the protein level (https://cdn.amegroups.cn/static/public/atm-22-4815-4.xlsx). The PPI network was used for further analysis in Cytoscape (Figure S1). The number of interactions proteins was calculated and sorted (Figure 3A). As shown in the bar plot, AKT1 was the most frequent protein that other proteins interacted with. Meanwhile, the compound-candidate genes network was visualized by Cytoscape. The more interactions between candidate genes and active ingredients, the darker and larger nodes would be. The network indicated that each active ingredient targeted multiple candidate genes, with PTGS1 and PTGS2, also known as cyclooxygenase 1 and 2 (COX-1, COX2), being the most frequently targeted genes (Figure 3B).
GO enrichment and KEGG enrichment analysis
Totals of 2,270 BP GO terms, 48 CC GO terms, and 143 MF GO terms were identified by the R package cluster profiler. The top 10 GO terms of each aspect were exhibited (Figure 4A). In terms of BP, the results showed that candidate genes play an important role in response to external stress and macromolecules. As for CC, candidate genes mainly affected proteins located on the cell membrane. The MF analysis indicated that protein binding capacity and enzyme receptor activity were regulated. The KEGG analysis showed that candidate genes participated in 167 KEGG signaling pathways. The top 10 KEGG pathways are listed in Table 1. In general, KEGG enrichment analysis indicated that candidate genes were involved in signaling pathways that pertained to immune functions such as the IL-17 signaling pathway (has04657), TNF signaling pathway (hsa04668), Th17 cell differentiation (hsa04659), and the T cell receptor signaling pathway (hsa04660). Besides, the PI3K-AKT signaling pathway (hsa04151) and MAPK signaling pathway (hsa04010) were two prevailing signaling pathways that candidate genes enriched (Figure 4B).
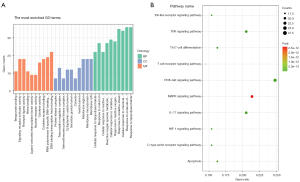
Table 1
| Pathway | Hub genes in pathway | Count | Adjp |
|---|---|---|---|
| IL-17 signaling pathway | MAPK14, RELA, JUN, FOS, MAPK1, IL6, NFKBIA | 24 | 2.89E-22 |
| TNF signaling pathway | MAPK14, RELA, JUN, AKT1, FOS, MAPK1, IL6, NFKBIA | 24 | 1.6E-20 |
| Th17 cell differentiation | MAPK14, RELA, JUN, FOS, MAPK1, IL6, NFKBIA, IL2 | 19 | 5.93E-15 |
| Toll-like receptor signaling pathway | MAPK14, RELA, JUN, AKT1, MAPK1, IL6, NFKBIA | 18 | 4.51E-14 |
| C-type lectin receptor signaling pathway | MAPK14, RELA, JUN, AKT1, MAPK1, IL6, NFKBIA, L2 | 18 | 4.51E-14 |
| PI3K-Akt signaling pathway | RELA, AKT1, MAPK1, IL6, IL2 | 29 | 7.58E-14 |
| HIF-1 signaling pathway | RELA, AKT1, MAPK1, IL6 | 18 | 9.16E-14 |
| T cell receptor signaling pathway | MAPK14, RELA, JUN, AKT1, FOS, MAPK1, NFKBIA | 17 | 5.73E-13 |
| Apoptosis | RELA, JUN, AKT1, FOS, MAPK1, NFKBIA | 19 | 3.54E-13 |
| MAPK signaling pathway | MAPK14, RELA, JUN, AKT1, FOS, MAPK1, MYC | 25 | 2.58E-12 |
Adjp, adjust P value. IL-17, interleukin 17; TNF, tumor necrosis factor; HIF, Hypoxia-Inducible Factor; MAPK, mitogen-activated protein kinase.
Hub gene screening
To screen the core genes among 121 candidate genes, the primary subnetwork and secondary subnetwork were screened based on the following parameters: Betweenness, Closeness, Degree, Eigenvector, LAC, and Network, and genes with values of all parameters that were higher than average were highlighted (Figure 5A,5B). In the end, 11 hub genes, NFKBIA, MAPK14, IL2, IL6, MAPK1, AKT1, ESR1, MYC, FOS, JUN, and RELA, were preserved after screening (Figure 5C). Function analyses were performed on the hub genes. GO analysis confirmed that the hub genes were closely related to inflammation and immune function (Figure S2A). Besides, KEGG analysis revealed that, through PI3K-AKT signaling pathway, the hub genes worked (Figure S2B).
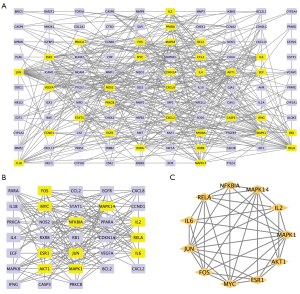
Molecular docking simulation
The potential interaction activity between 11 hub genes and their related HMM compounds was explored in this study using molecular docking analysis. Meanwhile, the docking affinity values supplied by AutoDock Vina were used to further select the obtained targets and active molecules. In total, 23 pairs were delivered to the docking simulation (Table 2). The stronger the binding ability between the compounds and the active sites of the targets, the higher the absolute value of the docking affinity. Most binding complexes had a high binding affinity, with an average of −8.73 kcal/mol, according to the docking studies. Among the complexes composed by active ingredients and hub genes, the top 10 complexes were selected based on binding affinity which were FOS-quercetin complex (−10.4 kcal/mol), AKT-kaempferol complex (−9.6 kcal/mol), RELA-kaempferol complex (−9.5 kcal/mol), AKT-quercetin complex (−9.4 kcal/mol), RELA-isorhamnetin complex (−9.2 kcal/mol), RELA-quercetin complex (−9.2 kcal/mol), JUN-kaempferol complex (−9.2 kcal/mol), JUN-quercetin complex (−8.8 kcal/mol), JUN-formononetin complex (−8.4 kcal/mol), and MAPK1-quercetin complex (−8.4 kcal/mol) (Figure 6).
Table 2
| Number | Hub genes | Compound | Docking affinity (kcal/mol) |
|---|---|---|---|
| 1 | AKT1 | Kaempferol | −9.6 |
| 2 | AKT1 | Quercetin | −9.4 |
| 3 | ESR1 | 3,9-di-O-methylnissolin | −6.9 |
| 4 | ESR1 | 7-O-methylisomucronulatol | −6.8 |
| 5 | ESR1 | Calycosin | −7.8 |
| 6 | ESR1 | Formononetin | −8.4 |
| 7 | ESR1 | Isorhamnetin | −8.0 |
| 8 | FOS | Quercetin | −10.4 |
| 9 | IL2 | Quercetin | −7.9 |
| 10 | IL6 | Quercetin | −7.8 |
| 11 | JUN | Formononetin | −8.4 |
| 12 | JUN | Kaempferol | −9.2 |
| 13 | JUN | Quercetin | −8.8 |
| 14 | MAPK1 | Quercetin | −8.4 |
| 15 | MAPK14 | 7-O-methylisomucronulatol | −7.0 |
| 16 | MAPK14 | Calycosin | −8.2 |
| 17 | MAPK14 | Formononetin | −7.8 |
| 18 | MAPK14 | Isorhamnetin | −8.5 |
| 19 | MYC | Quercetin | −7.6 |
| 20 | NFKBIA | Quercetin | −7.7 |
| 21 | RELA | Isorhamnetin | −9.2 |
| 22 | RELA | Kaempferol | −9.5 |
| 23 | RELA | Quercetin | −9.2 |
HMM, Hedysarum multijugum Maxim.
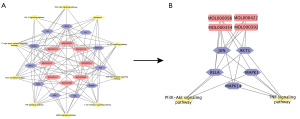
Compound-hub genes-signaling pathway network
After two rounds of screening, MOL000098 (quercetin, degree =9), MOL000422 (kaempferol, degree =3), MOL000392 (formononetin, degree =3), and MOL000354 (isorhamnetin, degree =3), were identified as the ingredients which had frequent interactions with hub genes, RELA (degree =12), MAPK14 (degree =11), MAPK1 (degree =10), JUN (degree =10), and AKT1 (degree =9) (Figure 7A). According to KEGG analysis, the PI3K-AKT and TNF signaling pathways were the signaling pathways with the highest involvement with these five hub genes. A critical network was then established based on these selected active ingredients, hub genes, and signaling pathways (Figure 7B). Therefore, the underlying mechanism by which HMM treats UC was shown intuitively. Additionally, the PI3K-AKT signaling pathway and TNF signaling pathway with enriched candidate genes were highlighted.
Discussion
Originally characterized in 1859, UC is one of two primary types of IBD (28). The incidence of UC is rising worldwide, with that of developed countries staying stable and developing countries increasing, especially India (29). The etiology of UC is not completely understood. Genetic background, environmental and internal factors, and immune dysregulation are regarded as possible risk factors (30). Currently, UC is diagnosed using a combination of clinical symptoms, endoscopic appearance, and histological outcomes. Bloody diarrhea is the most common symptom of UC. Endoscopic results usually show that the mucosa is consistently irritated and extending proximally from the anorectal verge, with the extent deteriorating from mild (only granular appearance) to severe UC (shallow ulcerations). Histological examination suggests that the inflammation is generally confined to the mucosal layer (1). In patients with UC, the therapeutic strategy is mostly determined by the severity of the illness. For acute severe UC patients, hospitalization is required, and intravenous corticosteroids and anticoagulants are applied (31). Medically refractory UC, poor medication tolerance, and UC-associated colorectal cancer are the most common reasons for surgery in UC patients (32). 5-aminosalicylic acid (5-ASA), oral corticosteroids, and thiopurines are the first-line treatment for mild-to-moderate patients. There is about a 50% clinical remission rate reported for 5-ASA treatments after two weeks. Thus, oral corticosteroids are the next step. However, remission and side effects cannot be prevented after oral corticosteroids. Besides, the use of thiopurines is limited because of the slow onset and severe adverse effects (33). Therefore, the efficiency of first-line treatment is unsatisfactory as expected. Nowadays, many hospitals regard TCM as a primary treatment, as the effect is surprisingly positive, and research focusing on TCM is mounting (5-7). In the present study, a TCM named Huangqi, also known as Hedysarum multijugum Maxim (HMM), is a common ingredient in plenty of decoctions, piqued our attention owing to the favorable outcomes in treating UC patients.
According to TCMSP, HMM is the root of the leguminous plant safflower astragalus. Based on previous studies, HMM has been shown to take effect on many diseases. Yang et al. indicated that HMM treated ischemic stroke and protected patients from cerebral ischemia-reperfusion injury by regulating inflammation, oxidative stress, endoplasmic reticulum stress, and angiogenesis through signaling pathways, such as the PI3K-AKT, FoxO, TNF, HIF-1, Rap1, and VEGF signaling pathways (34,35). Zhang et al. showed that HMM improved the general condition of diabetic cardiomyopathy in a rat model (36). Even for coronavirus disease of 2019 (COVID-19), HMM made its contribution to treating COVID-19 pneumonia by targeting PTGS2, TNF, and IL-6 and regulating the TNF and IL-17 signaling pathways (37). The important role HMM plays in treating UC has drawn much attention and has been researched in a UC rat model, yet no exact conclusion has been reached (8,9). Exploring the pathways of compounds, proteins, or genes is the goal of network pharmacology, which can help explain the complexity of biological systems, drugs, and diseases. Besides, the docking sites for active ingredients and key targets of drugs can be predicted by molecular docking. Thereby, novel drugs developed with specific ingredients greatly improve clinical efficacy and reduce toxicity and side effect. For example, the underlying mechanism of Liu Jun An Wei formula in treating gastrointestinal reactions caused by chemotherapy for colorectal cancer is explored by network pharmacology (38). Besides, He et al. revealed that Danshen promoted JAL-STAT signaling pathway to treat anemia according to network pharmacology (39). In addition, it is reported that Moluodan treats chronic atrophic gastritis by regulating NF-kB signaling pathway (40). Therefore, in this study, network pharmacology and molecular docking were applied to explore the possible mechanism behind HMM treating UC.
As shown above, 20 active ingredients and 121 candidate genes were screened. The gene PTGS2 is the most frequent target of active ingredients, followed by PTGS1 and other genes. Both of PTGS2 and PTGS1 encode COX2 and COX1 to form prostanoids which are essential molecules in the inflammatory response and pain (41). Singer et al. indicated that upregulated COX2 was detected in UC patients compared to healthy controls, providing a possible pathogenesis for UC (42). Therefore, by targeting COX2, HMM not only inhibits the inflammatory response but also restricts the development of UC in the first place. Whereas, as shown in PPI analysis, AKT1 had the most connections with other protein genes encoded by candidate genes. AKT1 is a serine/threonine-specific protein kinase activated by phosphoinositide 3-kinases (PI3K) which regulate a majority of biological processes, such as proliferation, migration, angiogenesis, metabolism, and inflammation (43). The detailed function of AKT1 in treating UC would be discussed in the next part. An abundance of research has already shown that AKT1 and COX2 cooperate to fulfill complicated missions. St-Germain et al. had proved that AKT regulated COX2 expression through the NF-kB pathway (44,45). Besides, according to Leng et al., COX2 positively regulates AKT phosphorylation (46). Therefore, it is reasonable to infer that HMM regulates top genes in bar plots, such as AKT and JUN, to inhibit the inflammatory response through COX2 and other frequent target candidate genes. Relying on research in recent years, the understanding of UC’s pathogenesis has improved. Impaired epithelial barrier and intestinal microbiota dysfunction initiate neutrophil extracellular traps (NET) and immune responses which produce various cytokines in lamina propria of intestine, which, in the end, evolve into inflammatory responses. Cytokines such as TNF, IL-9, IL-13, IL-23, and IL-36 are important immune mediators in UC pathogenesis (28). Therefore, UC can also be defined as an immune disease. The results of GO and KEGG analysis were consistent with the studies mentioned above and matched novel findings. In summary, HMM improves intestinal responses to external stress, such as nutrients, bacteria, and oxidative stress, which probably protects the intestine from epithelial damage and microbiota dysfunction, through the PI3K-AKT signaling pathway and some immune-related signaling pathways. To gain further understanding of HMM treating UC, 11 hub genes, all of which belong to the top 30 interacted genes, were preserved among candidate gene lists. Based on the compound-candidate gene network, eight active ingredients targeting hub genes showed up. A network comprising 11 hub genes, eight active ingredients, and the top 10 enriched signaling pathways was drawn, and a critical network was established after eliminating trivial elements, leaving two signaling pathways, four active ingredients, and five hub genes.
These four active ingredients all belong to the flavonols family which endows them with similar properties; they are known for their anti-inflammation and antioxidant effects (47-50), yet differences remain between them. Consistent with the former conclusion, it was reported that quercetin restricted COX2 expression by inhibiting PI3K phosphorylation, which consequently inhibited AKT1 phosphorylation and downstream gene expression (51,52). Apart from the PI3K signaling pathway, studies have shown that other pro-inflammation signaling pathways, MAPK, and JAK/STAT3, were also inhibited by kaempferol (53,54). According to recent research, quercetin attenuates intestinal damage and senescence by regulating increasing antioxidant capacity (55,56). Another important function of kaempferol is improving intestinal barrier function and integrity. By suppressing the activation of the NF-kB signaling pathway, kaempferol improved barrier function in a lipopolysaccharide (LPS)-induced epithelial-endothelial coculture model (57). Besides, quercetin has been widely applied in wound management owing to its migration and proliferation enhancing abilities (58). Thus, intestinal epithelial repair could be accelerated in UC patients treated with quercetin and kaempferol. As for formononetin, its primary function is antibacterial and antiviral (49). Research indicated that formononetin reshapes the intestinal microbiota by increasing maximum bacteria genera (59). Progestational hormone X receptor (PXR)-mediated up-regulation of xenobiotic metabolism and down-regulation of NF-kB signaling ameliorates the effects of isorhamnetin on experimental IBD. The findings might help to improve the use of isorhamnetin or its derivatives as a PXR ligand in the treatment of human IBD (60). Altogether, HMM treats UC through three aspects: diminishing inflammation, restoring intestinal homeostasis, and accelerating healing. Besides, studies have reported that quercetin and kaempferol inhibits TNF, IL9, and IL13 production (61,62). Thus, regulating immune responses is another pathway through which HMM treats UC.
Active ingredients function through regulating the hub genes mentioned above. According to Zhang et al., AKT was activated by fibrinogen and caused colitis by increasing vascular permeability in the mice model (63). However, post-translational modification, like SUMOylation, was inactivated in UC patients (64). Therefore, the form of AKT1 determines its role in UC: pAKT promotes UC development, and SUMOylated-AKT1 protects the intestine from colitis. Besides, degradation of AKT2 via ubiquitination reduced colonic damage in the TNBS-induced UC mice model (65). MAPK14, an isoform of serine/threonine-specific kinase (SAPKs) is a major subfamily of mitogen-activated protein kinase, which plays an important role in TNF-alpha production (66). Patients with mutated MAPK14 showed adverse therapeutic effects when treated with glucocorticoid, suggesting that MAPK14 is a potential biomarker to predict therapeutic responses (67). Research has found that UC patients with high MAPK14 expression experience more severe abdominal pain than patients with low MAPK14 expression (68). JUN, also known as a subunit of the AP-1 transcription factor, decreased as dextran sulfate sodium (DSS)-induced colitis mice condition improved (69). RELA encodes transcription factor p65 which is a subunit of nuclear factor NF-kB p65 subunit. Many researchers have studied the function of NF-kB in UC. In general, these two transcription factors are used to activate signaling pathways that HMM regulated. As for MAPK1, no study has focused on its relationship with UC. The results of molecular docking showed that active ingredients have a good binding affinity with receptor proteins encoded by these hub genes, especially AKT-kaempferol complex (−9.6 kcal/mol), RELA-kaempferol complex (−9.5 kcal/mol), and AKT-quercetin complex (−9.4 kcal/mol). Kaempferol and quercetin targeted hub genes with the highest efficiency, which indicated that derivatives or synthetic drugs processed from kaempferol and quercetin could be an advanced solution for UC.
Concerning signaling pathways, PI3K-AKT and TNF were selected as most hub genes enriched in these signaling pathways. The PI3K-AKT signaling pathway participates in various processes and dysregulation of this pathway leads to multiple diseases, such as cancer, inflammation, and immune diseases (70,71). Research has shown that inhibition of the PI3K-AKT signaling pathway relieves UC symptoms and histological damage in mice models owing to the reduction of pro-inflammatory cytokines such as TNF (72). The TNF-alpha signaling pathway is activated when TNF binds to its receptor on the cell membrane. The immune system is activated, either innate immune or adaptive immune, when TNF binds to TNF receptor 2 on immune cells like T cells, macrophages, and dendritic cells (73). According to studies, TNF is increased and related to intestinal barrier defects in UC patients (74). As pathophysiological mechanisms are explored, the emerging treatments vary. In recent years, anti-TNF therapies are becoming increasingly essential in UC treatment, such as infliximab, adalimumab, and golimumab (75). Hence, HMM inhibits PI3K-AKT signaling to reduce TNF production and inhibit the TNF signaling pathway, which functions similarly to anti-TNF therapy.
In summary, this study provides a theory that in the treatment of UC, the active ingredients of HMM, namely, quercetin, kaempferol, formononetin, and isorhamnetin regulate the PI3K-AKT and TNF signaling pathways by targeting essential genes, AKT1, JUN, MAPK14, RELA, and MAPK1. Still, the results need to be validated in vivo or in vitro. To a certain extent, the reliability of experimental results is affected by the inadequacies of the network pharmacology database. Hence, there is an urgent need to complete the construction of the database.
Conclusions
Overall, TCM provides novel UC treatments with high efficiency and security. In this study, five active ingredients and five hub genes were selected as key molecules in the treatment of UC by HMM through PPI analysis, GO and KEGG analyses, and molecular docking simulation. The PI3K-AKT and TNF signaling pathways were found to be the most targeted signaling pathways, which pointed out that the active ingredients control inflammation in the intestines through immune regulation and suppression. This study suggested that innovative drugs based on these active ingredients could be an efficient and safe way of treating UC. However, further experiments are needed validate our findings and promote the feasibility of this approach.
Acknowledgments
Funding: This work was supported by the Key Research and Development Program of Shandong Province (No. 2021CXGC011104; No. 2019JZZY010104; No. 2019GSF108146), the Academic Promotion Program of Shandong First Medical University (No. 2019QL021), and the Special Foundation for Taishan Scholars Program of Shandong Province (No. ts20190978).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4815/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4815/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med 2011;365:1713-25. [Crossref] [PubMed]
- Ordás I, Eckmann L, Talamini M, et al. Ulcerative colitis. Lancet 2012;380:1606-19. [Crossref] [PubMed]
- Rozich JJ, Holmer A, Singh S. Effect of Lifestyle Factors on Outcomes in Patients With Inflammatory Bowel Diseases. Am J Gastroenterol 2020;115:832-40. [Crossref] [PubMed]
- Feuerstein JD, Moss AC, Farraye FA. Ulcerative Colitis. Mayo Clin Proc 2019;94:1357-73. [Crossref] [PubMed]
- Wei M, Li H, Li Q, et al. Based on Network Pharmacology to Explore the Molecular Targets and Mechanisms of Gegen Qinlian Decoction for the Treatment of Ulcerative Colitis. Biomed Res Int 2020;2020:5217405. [Crossref] [PubMed]
- Zhu L, Gu P, Shen H. Protective effects of berberine hydrochloride on DSS-induced ulcerative colitis in rats. Int Immunopharmacol 2019;68:242-51. [Crossref] [PubMed]
- Huang C, Dong J, Jin X, et al. Intestinal anti-inflammatory effects of fuzi-ganjiang herb pair against DSS-induced ulcerative colitis in mice. J Ethnopharmacol 2020;261:112951. [Crossref] [PubMed]
- Zang K, Wu J, Duan H, et al. Effect and the underlying mechanism of astragaloside IV on ulcerative colitis in rats. The Chinese Journal of Clinical Pharmacology 2019;35:48-51.
- Hao L, Wu Y, Li Y, et al. Mechanism of astragaloside IV in the treatment of ulcerative colitis in rats. Journal of Chinese Practical Diagnosis and Therapy 2019;33:439-42.
- Hopkins AL. Network pharmacology. Nat Biotechnol 2007;25:1110-1. [Crossref] [PubMed]
- Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci 2019;20:4331. [Crossref] [PubMed]
- Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med 2013;11:110-20. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13:6964-82. [Crossref] [PubMed]
- Varma MV, Obach RS, Rotter C, et al. Physicochemical space for optimum oral bioavailability: contribution of human intestinal absorption and first-pass elimination. J Med Chem 2010;53:1098-108. [Crossref] [PubMed]
- UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506-15. [Crossref] [PubMed]
- Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: integrating information about genes, proteins and diseases. Trends Genet 1997;13:163. [Crossref] [PubMed]
- Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res 2002;30:163-5. [Crossref] [PubMed]
- Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res 2002;30:412-5. [Crossref] [PubMed]
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Missiuro PV, Liu K, Zou L, et al. Information flow analysis of interactome networks. PLoS Comput Biol 2009;5:e1000350. [Crossref] [PubMed]
- Raman K, Damaraju N, Joshi GK. The organisational structure of protein networks: revisiting the centrality-lethality hypothesis. Syst Synth Biol 2014;8:73-81. [Crossref] [PubMed]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2018;46:8-13. [Crossref] [PubMed]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235-42. [Crossref] [PubMed]
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785-91. [Crossref] [PubMed]
- Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers 2020;6:74. [Crossref] [PubMed]
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769-78. [Crossref] [PubMed]
- Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. Lancet 2017;389:1756-70. [Crossref] [PubMed]
- Van Assche G, Vermeire S, Rutgeerts P. Management of acute severe ulcerative colitis. Gut 2011;60:130-3. [Crossref] [PubMed]
- Nguyen GC, Bernstein CN, Bitton A, et al. Consensus statements on the risk, prevention, and treatment of venous thromboembolism in inflammatory bowel disease: Canadian Association of Gastroenterology. Gastroenterology 2014;146:835-848.e6. [Crossref] [PubMed]
- Harbord M, Eliakim R, Bettenworth D, et al. Third European Evidence-based Consensus on Diagnosis and Management of Ulcerative Colitis. Part 2: Current Management. J Crohns Colitis 2017;11:769-84. [Crossref] [PubMed]
- Yang K, Zeng L, Ge A, et al. The Effect of Hedysarum multijugum Maxim.-Chuanxiong rhizoma Compound on Ischemic Stroke: A Research Based on Network and Experimental Pharmacology. Oxid Med Cell Longev 2020;2020:6072380. [Crossref] [PubMed]
- Yang K, Zeng L, Ge A, et al. Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury's Biological Network Based on Systematic Pharmacology. Front Pharmacol 2021;12:601846. [Crossref] [PubMed]
- Zhang S, Yuan Z, Wu H, et al. Network Pharmacology-Based Strategy Reveals the Effects of Hedysarum multijugum Maxim.-Radix Salviae Compound on Oxidative Capacity and Cardiomyocyte Apoptosis in Rats with Diabetic Cardiomyopathy. Biomed Res Int 2020;2020:8260703. [Crossref] [PubMed]
- Ye M, Luo G, Ye D, et al. Network pharmacology, molecular docking integrated surface plasmon resonance technology reveals the mechanism of Toujie Quwen Granules against coronavirus disease 2019 pneumonia. Phytomedicine 2021;85:153401. [Crossref] [PubMed]
- Li G, Liu L, Yin Y, et al. Network pharmacology and experimental verification-based strategy to explore the underlying mechanism of Liu Jun An Wei formula in the treatment of gastrointestinal reactions caused by chemotherapy for colorectal cancer. Front Pharmacol 2022;13:999115. [Crossref] [PubMed]
- He S, Wang T, Shi C, et al. Network pharmacology-based approach to understand the effect and mechanism of Danshen against anemia. J Ethnopharmacol 2022;282:114615. [Crossref] [PubMed]
- Zhou W, Zhang H, Wang X, et al. Network pharmacology to unveil the mechanism of Moluodan in the treatment of chronic atrophic gastritis. Phytomedicine 2022;95:153837. [Crossref] [PubMed]
- Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem 2000;69:145-82. [Crossref] [PubMed]
- Singer II, Kawka DW, Schloemann S, et al. Cyclooxygenase 2 is induced in colonic epithelial cells in inflammatory bowel disease. Gastroenterology 1998;115:297-306. [Crossref] [PubMed]
- Thompson JE, Thompson CB. Putting the rap on Akt. J Clin Oncol 2004;22:4217-26. [Crossref] [PubMed]
- St-Germain ME, Gagnon V, Mathieu I, et al. Akt regulates COX-2 mRNA and protein expression in mutated-PTEN human endometrial cancer cells. Int J Oncol 2004;24:1311-24. [Crossref] [PubMed]
- St-Germain ME, Gagnon V, Parent S, et al. Regulation of COX-2 protein expression by Akt in endometrial cancer cells is mediated through NF-kappaB/IkappaB pathway. Mol Cancer 2004;3:7. [Crossref] [PubMed]
- Leng J, Han C, Demetris AJ, et al. Cyclooxygenase-2 promotes hepatocellular carcinoma cell growth through Akt activation: evidence for Akt inhibition in celecoxib-induced apoptosis. Hepatology 2003;38:756-68. [Crossref] [PubMed]
- Li Y, Yao J, Han C, et al. Quercetin, Inflammation and Immunity. Nutrients 2016;8:167. [Crossref] [PubMed]
- Devi KP, Malar DS, Nabavi SF, et al. Kaempferol and inflammation: From chemistry to medicine. Pharmacol Res 2015;99:1-10. [Crossref] [PubMed]
- Machado Dutra J, Espitia PJP, Andrade Batista R. Formononetin: Biological effects and uses - A review. Food Chem 2021;359:129975. [Crossref] [PubMed]
- Gong G, Guan YY, Zhang ZL, et al. Isorhamnetin: A review of pharmacological effects. Biomed Pharmacother 2020;128:110301. [Crossref] [PubMed]
- Lee KM, Hwang MK, Lee DE, et al. Protective effect of quercetin against arsenite-induced COX-2 expression by targeting PI3K in rat liver epithelial cells. J Agric Food Chem 2010;58:5815-20. [Crossref] [PubMed]
- Endale M, Park SC, Kim S, et al. Quercetin disrupts tyrosine-phosphorylated phosphatidylinositol 3-kinase and myeloid differentiation factor-88 association, and inhibits MAPK/AP-1 and IKK/NF-κB-induced inflammatory mediators production in RAW 264.7 cells. Immunobiology 2013;218:1452-67. [Crossref] [PubMed]
- Huang CH, Jan RL, Kuo CH, et al. Natural flavone kaempferol suppresses chemokines expression in human monocyte THP-1 cells through MAPK pathways. J Food Sci 2010;75:H254-9. [Crossref] [PubMed]
- Gong JH, Shin D, Han SY, et al. Blockade of Airway Inflammation by Kaempferol via Disturbing Tyk-STAT Signaling in Airway Epithelial Cells and in Asthmatic Mice. Evid Based Complement Alternat Med 2013;2013:250725. [Crossref] [PubMed]
- Saccon TD, Nagpal R, Yadav H, et al. Senolytic Combination of Dasatinib and Quercetin Alleviates Intestinal Senescence and Inflammation and Modulates the Gut Microbiome in Aged Mice. J Gerontol A Biol Sci Med Sci 2021;76:1895-905. [Crossref] [PubMed]
- Xu B, Qin W, Xu Y, et al. Dietary Quercetin Supplementation Attenuates Diarrhea and Intestinal Damage by Regulating Gut Microbiota in Weanling Piglets. Oxid Med Cell Longev 2021;2021:6221012. [Crossref] [PubMed]
- Bian Y, Dong Y, Sun J, et al. Protective Effect of Kaempferol on LPS-Induced Inflammation and Barrier Dysfunction in a Coculture Model of Intestinal Epithelial Cells and Intestinal Microvascular Endothelial Cells. J Agric Food Chem 2020;68:160-7. [Crossref] [PubMed]
- Chittasupho C, Manthaisong A, Okonogi S, et al. Effects of Quercetin and Curcumin Combination on Antibacterial, Antioxidant, In Vitro Wound Healing and Migration of Human Dermal Fibroblast Cells. Int J Mol Sci 2021;23:142. [Crossref] [PubMed]
- Naudhani M, Thakur K, Ni ZJ, et al. Formononetin reshapes the gut microbiota, prevents progression of obesity and improves host metabolism. Food Funct 2021;12:12303-24. [Crossref] [PubMed]
- Dou W, Zhang J, Li H, et al. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J Nutr Biochem 2014;25:923-33. [Crossref] [PubMed]
- Manjeet K R, Ghosh B. Quercetin inhibits LPS-induced nitric oxide and tumor necrosis factor-alpha production in murine macrophages. Int J Immunopharmacol 1999;21:435-43. [Crossref] [PubMed]
- Jia Z, Chen A, Wang C, et al. Amelioration effects of Kaempferol on immune response following chronic intermittent cold-stress. Res Vet Sci 2019;125:390-6. [Crossref] [PubMed]
- Zhang C, Chen H, He Q, et al. Fibrinogen/AKT/Microfilament Axis Promotes Colitis by Enhancing Vascular Permeability. Cell Mol Gastroenterol Hepatol 2021;11:683-96. [Crossref] [PubMed]
- Mustfa SA, Singh M, Suhail A, et al. SUMOylation pathway alteration coupled with downregulation of SUMO E2 enzyme at mucosal epithelium modulates inflammation in inflammatory bowel disease. Open Biol 2017;7:170024. [Crossref] [PubMed]
- Zhu Y, Shi Y, Ke X, et al. RNF8 induces autophagy and reduces inflammation by promoting AKT degradation via ubiquitination in ulcerative colitis mice. J Biochem 2020;168:445-53. [Crossref] [PubMed]
- Han J, Lee JD, Bibbs L, et al. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994;265:808-11. [Crossref] [PubMed]
- Skrzypczak-Zielinska M, Gabryel M, Marszalek D, et al. NGS study of glucocorticoid response genes in inflammatory bowel disease patients. Arch Med Sci 2021;17:417-33. [Crossref] [PubMed]
- Grossi V, Hyams JS, Glidden NC, et al. Characterizing Clinical Features and Creating a Gene Expression Profile Associated With Pain Burden in Children With Inflammatory Bowel Disease. Inflamm Bowel Dis 2020;26:1283-90. [Crossref] [PubMed]
- Kim TW, Shin JS, Chung KS, et al. Anti-Inflammatory Mechanisms of Koreanaside A, a Lignan Isolated from the Flower of Forsythia koreana, against LPS-Induced Macrophage Activation and DSS-Induced Colitis Mice: The Crucial Role of AP-1, NF-κB, and JAK/STAT Signaling. Cells 2019;8:1163. [Crossref]
- Patel RK, Mohan C. PI3K/AKT signaling and systemic autoimmunity. Immunol Res 2005;31:47-55. [Crossref] [PubMed]
- Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer 2020;20:74-88. [Crossref] [PubMed]
- Huang XL, Xu J, Zhang XH, et al. PI3K/Akt signaling pathway is involved in the pathogenesis of ulcerative colitis. Inflamm Res 2011;60:727-34. [Crossref] [PubMed]
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science 2002;296:1634-5. [Crossref] [PubMed]
- Lissner D, Schumann M, Batra A, et al. Monocyte and M1 Macrophage-induced Barrier Defect Contributes to Chronic Intestinal Inflammation in IBD. Inflamm Bowel Dis 2015;21:1297-305. [Crossref] [PubMed]
- Rutgeerts P, Sandborn WJ, Feagan BG, et al. Infliximab for induction and maintenance therapy for ulcerative colitis. N Engl J Med 2005;353:2462-76. [Crossref] [PubMed]


