
The downregulation of LINC00273 inhibits the proliferation, invasion, and migration of ovarian cancer cells in vivo and in vitro
Introduction
Ovarian cancer is a malignant tumor and is associated with increased mortality in women (1). Ovarian cancer is the 3rd most common malignant tumor related to the female reproductive system and is the most common cause of cancer-related death (1). Due to a lack of effective ovarian cancer screening strategies, the clinical symptoms are only identified late, resulting in widespread pelvic and abdominal implant dissemination. More than 60% of patients suffering from this type of cancer are in the terminal stage at the time of diagnosis (2). Currently, early and effective diagnostic strategies for ovarian cancer are still lacking. Due to their advanced stage at the time of diagnosis, patients have a poor prognosis. Ovarian cancer is widely treated with chemotherapy, surgery, gene therapy, and biological therapy; however, the 5-year survival rate of these patients is still only 35–38% (3,4). Thus, it is crucial that research be conducted on ovarian cancer to enable its early evaluation, diagnosis, and treatment.
A complex mechanism, which includes proto-oncogene activation, tumor suppressor gene inactivation, and other molecular alterations, underlies the incidence and development of ovarian cancer (4,5). For example, more and more long non-coding ribonucleic acids (lncRNAs) have been found to be associated with the biological mechanisms of these cancerous cells, including their migration, growth, invasion, apoptosis, and ageing (6,7). LncRNA is a newly discovered non-coding RNA molecule >200 nucleotides in length. It regulates gene expression by various means, such as transcriptional activation, chromatin modification, intranuclear transport, and transcriptional interference. For example, lncRNA BLACAT1 has been shown to inhibit the proliferation and invasion ability of ovarian cancer by miR-519-3p regulation and Wnt/β-catenin signaling pathway inhibition (8). Additionally, LncRNA MEG3 has been shown to suppress cellular proliferation, promote apoptosis, and inhibit cell cycle progression by modulating the downstream gene phosphate and tension homology deleted on chromsome ten (PTEN) associated with ovarian cancer cells (9). Further, lncRNA has also been shown to modulate ovarian cancer-related drug resistance. For example, in ovarian cancer drug-resistant cells, the expression of lncRNA HOTAIR is high; however, its downregulation can increase apoptosis and inhibit cell proliferation in drug-resistant ovarian cancer cells thereby inhibiting tumor development (10). These results show that lncRNAs are crucial elements related to the development, incidence, and drug resistance of ovarian cancer.
LINC00273 is a recently discovered class of non-coding RNA with a number of critical roles. Studies have shown that abnormal LINC00273 expression is associated with a variety of human cancers, and LINC00273 has been proposed as a tumor oncogene gene. For example, the predictive screening of oncogenic signaling pathways showed that LINC00273 is a crucial biomarker that could predict the prognosis of oral squamous cell carcinoma patients (11). The study also revealed that transforming growth factor beta mediates LINC00273 expression in lung cancer, which in turn activates zinc finger E-box binding homeobox 1 (ZEB1) by upregulating miR200a-3p to promote invasion and metastasis (12). The pyruvate dehydrogenase A1 binds to pyruvate kinase M2 to induce the expression of LINC00273 through histone acetylation (13). These results show that LINC00273 is an important molecule involved in the cell signaling pathways exerts biological functions, and may be a therapeutic target for tumors.
At present, previous research has shown the importance of LINC00273 in various tumors; however, research on the function of LINC00273 in malignant tumors is still limited and one-sided, and the role of LINC00273 in various solid tumors has not been widely described. LINC00273’s biological role in this type of cancer is still undetermined. This study sought to examine the expression of LINC00273 in ovarian cancerous tissues and ovarian cancerous cell lines, to observe its effect on the biological function of ovarian cancerous cells, and to preliminarily evaluate its molecular mechanism of action. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4562/rc).
Methods
Clinical sample collection
Ovarian cancer samples (cancerous and adjacent tissues) were taken from gynecological oncology surgery patients at the Yancheng First Hospital, Affiliated Hospital of Nanjing University Medical School. The ethical standards and related laws require that all patients are treated anonymously. None of the patients from whom the specimen sources were obtained underwent radiotherapy or chemotherapy before surgery. The histopathological type and tumor grade of the ovarian cancer was determined based on the pathological results by experts in the Pathology Department at The Second Affiliated Hospital of Soochow University, and the tumor stage was determined using the International Federation of Gynecology and Obstetrics (FIGO) staging method. The ovarian cancer surgical specimens were collected during the operation process. After the tumor specimens were removed, the cancer tissues and adjacent tissues were rapidly excised. After collection, they were quickly put into a RNA store sample preservation solution. The samples were soon transferred to −196 ℃ liquid nitrogen for 24 hours and then placed in a −80 ℃ refrigerator for long-term storage. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Yancheng First Hospital, Affiliated Hospital of Nanjing University Medical School (No. 2021-K012) and informed consent was taken from all the patients.
Cell culture
Normal human ovarian epithelial cells (IOSE80) and ovarian cancerous cell lines (OVCAR3, A2780, PEO1, and SKOV3) were acquired from the Shanghai Cell Bank, Chinese Academy of Sciences. IOSE80, A2780, and PEO1 were inoculated with Dulbecco’s Modified Eagle Medium (DMEM) to which 10% fetal bovine serum (FBS) was added. SKOV3 and OVCAR3 were inoculated in RPMI-1640 media supplemented with 10% FBS and cultured in a humidified incubator in a 5% carbon dioxide (CO2), 37 ℃ incubator. The log-growth phase cells were grown in 6-well plates, and upon reaching 50% cell density, the cells were transfected by lentivirus transfection with a plasmid containing the LINC00273 knockdown sequence or a negative control plasmid. Next, 6 h post-transfection, the exhausted media was removed, and fresh media was added. The human ovarian cancer cells are adherent growth cells, and the cells were cultured in a 5% CO2, 37 ℃ incubator. The culturing media contained 10% FBS and 1% penicillin-streptomycin. The cells were grown into monolayers in culture flasks, and the cells’ growth status was observed every day with an inverted microscope.
RNA extraction and qRT-PCR
The ovarian cancer tissues’ or cell lines’ total RNA was isolated using TRIzol, after which its concentration and purity was detected. Complementary deoxyribonucleic acid (DNA) was prepared by the reverse transcription of the RNA and reverse transcribe mRNA into cDNA. The expression of LINC00273 was detected with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference. The upstream GAPDH primer was 5'-ACAACTTTGG-TATCGTGGAAGG-3', and the downstream primer was 5'-GCCATCAC-GCCACAGTTTC-3. The primer of LINC00273 refer to this reference (14). The quantitative real-time polymerase chain reaction (qRT-PCR) parameters were pre-denaturation for 10 min at 94 ℃, for 30 s at 62 ℃, and 30 s at 72 ℃, for a total of 40 cycles. The gathered data were measured using the 2⁃ΔΔCt approach.
Detection of cell proliferation by Cell Counting Kit-8 (CCK8) assay
The CCK8 was used to detect the proliferative function of the human ovarian cancerous cells. The transfected cancerous ovarian cells were grown in 96-well plates, and 3 duplicate wells were made per group. Each well was inoculated with cell suspension (200 µL), whice was grown in a cell incubator overnight. CCK-8 reagent (20 µL) was inoculated at 0, 24, 48, and 72 h in each well, and the plate was then put in a CO2 incubator for 2 h. Finally, the absorbance value [Optical Density (OD) value] at 490 nm was recorded using a microplate reader, and cell proliferation was evaluated by analyzing the absorbance value.
Clonogenic assay
The clonogenic assay was conducted detect the LINC00273 knockdown effect on the cell clone formation ability. For details of the specific method used, refer to this previous publication (15).
EdU assay
The transfected ovarian cancerous cells were grown in a 24-well plate, EdU was supplemented to a final makeup volume of 10 µM, and the plate was left to incubate for 2 hours. Then the culturing media was aspirated, and 1 mL of fixative was used for fixation at room temperature (RT) for 15 min. The cells were then washed 3 times, for 5 mins each time, with 1 mL of washing solution, and 1 mL of permeabilization solution per well. The cells were then incubated for 10 min at RT. Next, 1 mL of Hoechst 33342 solution was added to each well, and the cells were then left to incubate in the dark at RT for 10 min. Next, the cells were washed with washing solution 3 times, for 3 min each time, and fluorescence detection was then performed.
Cell cycle detection by flow cytometry
Flow cytometry was performed to detect the cell cycles using a PI cell cycle detection kit. The ovarian cancer cells were collected, added to 1 mL of ice-bath pre-cooled 70% ethanol, and fixed at 4 ℃ for 12 h. Next, around 1 mL of ice-cold phosphate buffered solution (PBS) was added, and the solution was centrifuged to precipitate the cells. Pour off the supernatant carefully until 50 µL of PBS remains in the cell sample tube. Next, 0.5 mL of PI staining solution was added to each cell sample tube, and the cells were gradually and completely reconstituted. The cell pellets were suspended and incubated for 30 min in the dark at 37 ℃. The excitation wavelength of red fluorescence was detected at 488 nm in a flow cytometer, and, at the same time, light scattering was also detected. Finally, the cells’ DNA content was analyzed.
Transwell assay
A Matrigel invasion assay was performed to detect the cell invading capability of cells using a Matrigel gel-coated microporous membrane in the upper Transwell chamber. After the cells were re-suspended in FBS-free media, they were cultured in the upper Transwell chamber, at 200 µL media per well with a cell density of 5×104 cells/mL. In the lower chamber, 600 µL of DMEM media consisting of 10% FBS was added. After 24 h of culture, the surface attached cells on the microporous membrane upper chamber were gently wiped off with cotton swabs, and fixed for 30 min in paraformaldehyde at RT, washed 3 times with PBS, stained using 1% staining solution of crystal violet at RT for 30 min, and washed again with PBS solution 3 times. After drying at RT, the transmembrane cell number was selected randomly in 4 fields of view under a high-power microscope, photographed, and counted. The experiment was repeated 4 times.
Western blot detection
The cells in the log phase of growth were lysed to prepare the cell lysate. The total protein of each cell group was extracted from this lysate. After determining the protein amount, sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out using a protein concentration of 40 µg/well, and the electrophoretically isolated proteins were then transferred to the poly(vinylidene fluoride) (PVDF) membrane. The membrane was then blocked with 5% skim milk at RT for 2 h, incubated with the primary antibody at 4 ℃ overnight, and then, after washing the membrane, incubated with the secondary antibody at RT for 2 h. After another wash, development and exposure were performed using an efficient chemiluminescence (ECL) developer. GAPDH was used as the internal reference protein.
Nude mice subcutaneous tumorigenesis experiment
A total of 15 female BALB/c nude mice (aged 4–6 weeks old and weighing 18–20 g) were acquired from Nanjing University. The nude mice were randomly allocated to 3 groups, of 5 mice per group, based on their body weight, and reared in an Specific pathogen Free (SPF) environment. To ensure that the cells were in good condition, cells in each group were subjected to routine trypsin digestion, centrifugation, re-suspension, cell counting, and adjusting the density to 1×108/mL (re-suspended in PBS or serum-free medium). Nude mice were inoculated subcutaneously with 100 µL of cell suspension; that is, 1×107 cells (under the axilla of the right upper limb, with abundant blood supply).
Usually, a lump began to appear under the skin about a week after the inoculation. To evaluate the width (W) and length (L) of the subcutaneous tumor in the nude mice, vernier callipers were used once every 5 days. The tumor volume was measured. The following formula was used: tumor volume = 0.5× L × W2. Once the tumor grew to a certain size, the nude mice underwent uniform euthanasia, and the subcutaneously implanted tumor was extracted, measured, and photographed as soon as possible. Total RNA was extracted from tumor-bearing tissues.
Animal experiments were performed under a project license (No. IACUC-D2202118) granted by the Laboratory Animal Welfare Ethical Review Committee of Nanjing University, in compliance with Chinese guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Statistical analysis
SPSS 21.0 software was used for the statistical analysis. The results are presented as the mean ± standard deviation, and a t-test was used to compare the means among the groups. A P value <0.05 was considered statistically significant.
Results
Ovarian cancer tissues and cells highly express LINC00273
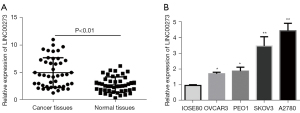
To further verify LINC00273 function in ovarian cancer, we used qRT-PCR to detect the LINC00273 expression in 30 ovarian cancerous and adjacent tissues along with ovarian cancer cell lines. The results revealed that compared to the normal adjacent tissues, the ovarian cancer tissues had high expression of LINC00273 (see Figure 1A). Further, in the normal human ovarian epithelial cells (IOSE80) and ovarian cancer cell lines (SKOV3, A2780, PEO1, and OVCAR3), the detection data indicated that in the ovarian cancer cell lines LINC00273 expression was markedly upregulated (see Figure 1B), indicating that LINC00273 may increase the incidence and development of ovarian cancer.
Establishment of an ovarian cancer cell line with stable knockdown of LINC00273
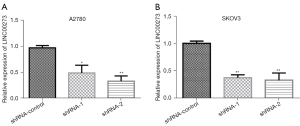
To further explore the distinct biological function of LINC00273 in ovarian cancer cells, we constructed lentiviral vectors [short hairpin RNA-1 (shRNA-1) and shRNA-2] and a blank control vector (shRNA-control) targeting the LINC00273 sequence. Transfection was then performed on the ovarian cancer cells A2780 and SKOV3, after which the LINC00273 expression level in the cells was detected by fluorescence quantitative PCR. The results showed that compared to the shRNA-control, the lentiviral vector shRNA-1 and shRNA-2 significantly downregulated LINC00273 expression in the ovarian cancerous cells (see Figure 2A,2B).
Effects of knockdown LINC00273 on the proliferative capability of ovarian cancerous cells
First, we detected the knockdown effect of LINC00273 on the proliferation of the cancerous ovarian cells A2780 and SKOV3. The cells were then transfected with shRNA-1, shRNA-2, or shRNA-NC and inoculated into 96-well plates 48 h post-transfection. Next, cell viability was detected by CCK-8 assay after 0, 24, 48, and 72 h of culture (see Figure 3A,3B). Compared to the shRNA-group cells, shRNA-1 and shRNA-2 had a significant effect on A2780 and SKOV3 cells at 24 h. However, cell proliferation was not largely affected. With the extension of the culture time to 48 h, the cells’ proliferation ability was greatly inhibited (P<0.05), and the inhibition effect was particularly significant at 72 h of culture (P<0.01).
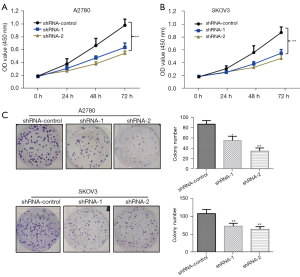
The clonogenic assay result indicated that compared to the shRNA-control, the knockdown of LINC00273 significantly suppressed the clonogenic ability of the A2780 ovarian cancerous cells. The knockdown of LINC00273 also significantly inhibited the ovarian cancerous cells’ colony formation capability (see Figure 3C). The above data indicate that the knockdown of LINC00273 inhibited the clonogenic capability of the ovarian cancerous cells. These experiments provide preliminary evidence that LINC00273 knockdown markedly inhibits A2780 and SKOV3 cell proliferation.
Effects of the knockdown of LINC00273 on ovarian cancer cell proliferation
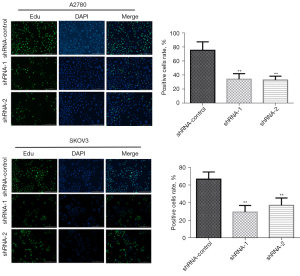
EdU is a thymidine analog that has the ability to replace Thymine (T) to infiltrate the replicating DNA molecule during cell proliferation. The replication activity of DNA was analyzed by a specific EdU and Apollo® fluorescent dye reaction that allows the accurate detection of EdU labeling, which reflects the proliferation of cells. The data indicated that in the A2780 cells, the knockdown of LINC00273 inhibited the DNA synthesis capability of the ovarian cancer cells more than the shRNA-control. In the SKOV3 cells, compared to the shRNA-control, the knockdown of LINC00273 also greatly reduced the DNA synthesis ability of the ovarian cancerous cells (see Figure 4). Our findings suggest that LINC00273 knockdown inhibited the DNA synthesis ability of ovarian cancerous cells, thereby inhibiting cell division and proliferation.
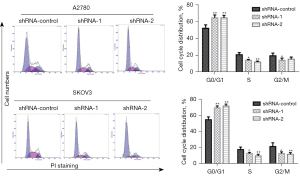
Effects of the knockdown of LINC00273 on the cell cycle of ovarian cancer cells
The above findings suggested that the knockdown of LINC00273 inhibited the DNA synthesis of the ovarian cancerous cells. The distribution of the knockdown of LINC00273 on the cell cycle was analyzed using the flow-cytometry protocol. In the A2780 cells, compared to the shRNA-control group, the knockdown LINC00273 highly promoted the ratio of ovarian cancerous cells in the G0/G1 phase and reduced the S and G2/M phase ratio. In the SKOV3 cells, compared to the shRNA-control group, the knockdown of LINC00273 also significantly promoted the ratio of the G0/G1 phase ovarian cancer cells and reduced the S and G2/M phase ratio (see Figure 5). These findings indicate that LINC00273 knockdown inhibits the ovarian cancer cell cycle.
Effects of the knockdown of LINC00273 on the migration and invasion of ovarian cancer cells
Numerous clinical cases have shown that cancer cells in individuals with ovarian cancer penetrate the ovarian capsule or fall off exogenous cancer cells scattered in the pelvis, resulting the deterioration of the patient’s condition. Thus, this study further evaluated the LINC00273 knockdown effect on the migration and invasion capability of ovarian cancer cells by Transwell assay. Compared to the shRNA-control group, knockdown of LINC00273 inhibited the migration of the A2780 ovarian cancerous cells in the Transwell plate from the upper to the lower chamber. LINC00273 also greatly inhibited the ovarian cancer cells’ ability to invade through Matrigel in the upper chamber and into the lower chamber. A similar phenomenon was also observed in the SKOV3 cells; that is, the knockdown of LINC00273 significantly inhibited the ovarian cancerous cells’ migration in the Transwell plate from the upper chamber to the lower chamber compared to the shRNA-control group. After the addition of Matrigel, the knockdown of LINC00273 also greatly suppressed ovarian cancer cells’ ability to invade through the upper Matrigel chamber into the lower chamber (see Figure 6). These results indicate that the knockdown of LINC00273 inhibited ovarian cancer cells’ capability to migrate and invade.
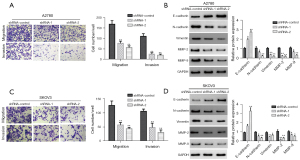
The LINC00273 knockdown effect on the migration and invasion-related protein expression of these cancerous cells was further evaluated by Western blot. The results showed that compared to the shRNA-control group, the knockdown of LINC00273 greatly promotes intracellular E-type cohesion (E-cadherin) expression and inhibits N-type cohesion (N-cadherin) and vimentin expression in the A2780 ovarian cancer cells. Further, the LINC00273 knockdown markedly inhibited matrix metalloproteinase-2 (MMP-2), and matrix metalloproteinase-9 (MMP-9) expression in ovarian cancerous cells (see Figure 6B). In the SKOV3 cells, a similar phenomenon was observed; that is, the knockdown of LINC00273 significantly promoted E-cadherin expression in the ovarian cancer cells and inhibited vimentin, MMP-2, N-cadherin, and MMP-9 expression (see Figure 6D).
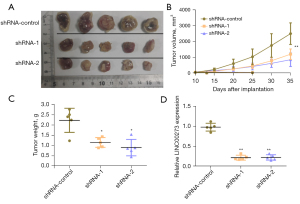
Effects of the knockdown of LINC00273 on the proliferation ability of ovarian cancer cells in vivo
The LINC00273 knockdown effect on the capability of ovarian cancerous cells to migrate and invade in vivo was further tested by in vivo tumorigenic experiments in nude mice. The data obtained indicated that compared to the shRNA-control group, the LINC00273 knockdown markedly inhibited the proliferative ability of these cancerous cells in nude mice subcutaneously (see Figure 7A,7B). The weighted analysis of the tumor-bearing tissues also showed that the weight of tumor-bearing tissues formed after the knockdown of LINC00273 was also greatly reduced compared to that of the shRNA-control group (see Figure 7C). The qRT-PCR analysis showed that the LINC00273 level of expression in the tumor-bearing tissues in the knockdown LINC00273 group was greatly reduced compared to that of the shRNA-control group (see Figure 7D).
Discussion
Previous investigation has reported that the occurrence and development ovarian cancer is a multi-stage process that includes apoptosis, invasion proliferation, migration, angiogenesis, and metastasis (16). Thus, in-depth knowledge of the molecular process involved in ovarian cancer pathogenesis is crucial to the prevention, diagnosis, and treatment of ovarian cancer.
Recent research has shown that a variety of lncRNAs are involved in the incidence and development of ovarian cancer (6). The present study found that LINC00273 was highly expressed in ovarian cancerous tissues. Our functional experiments showed that the knockdown of LINC00273 greatly inhibited the proliferation, and expression of vimentin, MMP-2 N-cadherin, and MMP-9 in ovarian cancer cells by promoting the E-cadherin expression. Additionally, in vivo experiments showed that knockdown of LINC00273 markedly suppressed the in vivo proliferation of ovarian cancer cells.
Previous research suggests that LINC00273 is a group of intracellular non-coding RNAs that have an important commonality and are crucial components involved in the incidence and development of various tumors, such as liver and lung cancer, and melanoma tumors (13,14,17). For example, the overexpression of LINC00273 promotes melanoma cancer metastasis, while LINC00273 knockdown greatly suppresses the proliferative function of tumor cells in vivo and in vitro (17). In lung cancer exosomes, Neural precursor cell-expressed developmentally downregulated 4 (NEDD4) is recruited by LINC00273 to promote the ubiquitination and degradation of large tumor suppressor kinase 2 (LATS2) to inactivate the Hippo pathway, and yes-associated protein (YAP) induces X-linked RNA-binding motif protein (RBMX) transcription to promote tumor metastasis and drug resistance (14). We found that the expression of LINC00273 was enhanced in the ovarian cancer tissues. The functional experiments suggested that the LINC00273 knockdown greatly reduced the proliferation of cancerous ovarian cells.
Metastasis is the primary reason behind the death of cancer patients and studying the molecular mechanisms that regulate the progression of cancer metastasis may provide some functional insights that can be used to establish effective diagnostic and therapeutic strategies (18). Previous research has shown that lncRNAs are important components in cancer metastasis (19,20). The literature indicates that abnormal lncRNA expression is often associated with ovarian cancer and can markedly affect the incidence and development of tumors. For example, lncRNA MSC-AS1 is under expressed in the cancerous tissues and cells of ovaries, and lncRNA MSC-AS1 overexpression suppresses cell growth and promotes apoptosis. Additionally, lncRNA MSC-AS1 targets and downregulates miR-425-5p (21). These results may lead to the identification of potential treatment targets for ovarian cancer.
The lncRNA CDKN2BAS is highly expressed in cancerous ovarian tissues and is associated with an advanced FIGO stage and pathological grade in patients. LncRNA CDKN2BAS overexpression enhances the proliferation and migration potential of ovarian cancer cells, while GAS6 knockdown partially abolishes the modulatory effect of lncRNA CDKN2BAS in promoting ovarian cancer proliferation and migration potential (22). These studies showed that lncRNAs have important application value in the incidence and establishment of ovarian cancer and disease diagnosis. Thus, it is crucial and that new lncRNAs with important functions are urgently identified.
Research has shown that lncRNA MALAT-1 induces the development and epithelial metastasis of ovarian cancer by sponging miRNA-22 (23). Additionally, lncRNA SNHG17 induces the proliferation and invasion ability of ovarian tumor cells by FOXA1 upregulation, which might be a potential ovarian cancer therapeutic target (24). Further, lncRNA TP73-AS1 induces cancer cell growth and metastasis in ovaries by MMP-2 and MMP-9 regulation (25). The findings of the present study suggest that the knockdown of LINC00273 significantly inhibits ovarian cancer cells’ invasion and migration ability by promoting the E-cadherin expression in these cells and inhibiting vimentin, MMP-2, N-cadherin, and MMP-9 expressions.
Additionally, lncRNAs can also act as important biomarkers for early diagnosis and be used to monitor the disease progression of ovarian cancer patients. For example, the lncRNA regulator of reprogramming (lncRNA ROR) expression in ovarian cancer patients’ plasma was significantly enhanced compared to that of normal subjects. Compared to para-cancerous normal tissue plasma, the lncRNA ROR level of ovarian cancer tissues has also been shown to be greatly increased. Further, studies have shown that lncRNA ROR expression is closely linked to FIGO stage, lymph node metastasis, and tumor grade. A Pearson correlation analysis revealed a positive correlation between lncRNA ROR and CA125 in ovarian cancer patients. The united evaluation of plasma lncRNA ROR and CA125 has a higher diagnostic value for ovarian cancer than the detection of lncRNA ROR or CA125 alone (26). Accumulating evidence indicates that some lncRNA play very active roles in OC progression, chemoresistance and recurrence through different molecular mechanisms (27-29). For example, LncRNA PVT1 regulated the transforming growth factor-β (TGF-β) pathway to promote ovarian cancer progression through sponging miR-148a-3p (30). LncRNA TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer through enhancing TLR8 mRNA stability (31). Thus, follow-up studies need to be conducted to explore whether the expression level of LINC00273 can be used as a possible biomarker for evaluating the metastasis of ovarian cancer patients.
In conclusion, LINC00273 is highly expressed in the cancerous tissues and cancerous cell lines of ovaries. The knockdown of LINC00273 greatly reduces the proliferation and invasion capability of these cancerous cells. The expression of cadherin, vimentin, MMP-2, and MMP-9 were also inhibited. Thus, LINC00273 can be considered a crucial target for the future treatment and prevention of ovarian cancer.
Acknowledgments
Funding: This study was supported by the State Key Laboratory of Soochow University (No. GZK1202106) and the Second Affiliated Hospital of Soochow University Superiority discipline Lifting Project (No. XKTJ-XK202006).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4562/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4562/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4562/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Yancheng First Hospital, Affiliated Hospital of Nanjing University Medical School (No. 2021-K012) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. IACUC-D2202118) granted by the Laboratory Animal Welfare Ethical Review Committee of Nanjing University, in compliance with Chinese guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yoshida R. Hereditary breast and ovarian cancer (HBOC): review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer 2021;28:1167-80. [Crossref] [PubMed]
- Vanacker H, Harter P, Labidi-Galy SI, et al. PARP-inhibitors in epithelial ovarian cancer: Actual positioning and future expectations. Cancer Treat Rev. 2021;99:102255. [Crossref] [PubMed]
- Wang Z, Meng F, Zhong Z. Emerging targeted drug delivery strategies toward ovarian cancer. Adv Drug Deliv Rev 2021;178:113969. [Crossref] [PubMed]
- Xie W, Sun H, Li X, et al. Ovarian cancer: epigenetics, drug resistance, and progression. Cancer Cell Int 2021;21:434. [Crossref] [PubMed]
- Kurnit KC, Fleming GF, Lengyel E. Updates and New Options in Advanced Epithelial Ovarian Cancer Treatment. Obstet Gynecol 2021;137:108-21. [Crossref] [PubMed]
- Braga EA, Fridman MV, Moscovtsev AA, et al. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int J Mol Sci 2020;21:8855. [Crossref] [PubMed]
- Zhan L, Li J, Wei B. Long non-coding RNAs in ovarian cancer. J Exp Clin Cancer Res 2018;37:120. [Crossref] [PubMed]
- Yang H, Qi Y, Wang XL, et al. Down-regulation of lncRNA BLACAT1 inhibits ovarian cancer progression by suppressing the Wnt/β-catenin signaling pathway via regulating miR-519d-3p. Mol Cell Biochem 2020;467:95-105. [Crossref] [PubMed]
- Wang J, Xu W, He Y, et al. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res 2018;67:927-36. [Crossref] [PubMed]
- Liu S, Lei H, Luo F, et al. The effect of lncRNA HOTAIR on chemoresistance of ovarian cancer through regulation of HOXA7. Biol Chem 2018;399:485-97. [Crossref] [PubMed]
- Fan WL, Yang LY, Hsieh JC, et al. Prognostic Genetic Biomarkers Based on Oncogenic Signaling Pathways for Outcome Prediction in Patients with Oral Cavity Squamous Cell Carcinoma. Cancers (Basel) 2021;13:2709. [Crossref] [PubMed]
- Sarkar A, Rahaman A, Biswas I, et al. TGFβ mediated LINC00273 upregulation sponges mir200a-3p and promotes invasion and metastasis by activating ZEB1. J Cell Physiol 2020;235:7159-72. [Crossref] [PubMed]
- Hossain AJ, Islam R, Kim JG, et al. Pyruvate Dehydrogenase A1 Phosphorylated by Insulin Associates with Pyruvate Kinase M2 and Induces LINC00273 through Histone Acetylation. Biomedicines 2022;10:1256. [Crossref] [PubMed]
- Chen J, Zhang K, Zhi Y, et al. Tumor-derived exosomal miR-19b-3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin Transl Med 2021;11:e478. [Crossref] [PubMed]
- Li W, Xu S, Peng N, et al. Downregulation of METTL7B Inhibits Proliferation of Human Clear Cell Renal Cancer Cells In Vivo and In Vitro. Front Oncol 2021;11:634542. [Crossref] [PubMed]
- Motohara T, Katabuchi H. Ovarian Cancer Stemness: Biological and Clinical Implications for Metastasis and Chemotherapy Resistance. Cancers (Basel) 2019;11:907. [Crossref] [PubMed]
- Jana S, Jana J, Patra K, et al. LINCRNA00273 promotes cancer metastasis and its G-Quadruplex promoter can serve as a novel target to inhibit cancer invasiveness. Oncotarget 2017;8:110234-56. [Crossref] [PubMed]
- Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci 2019;20:2767. [Crossref] [PubMed]
- Fares J, Fares MY, Khachfe HH, et al. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther 2020;5:28. [Crossref] [PubMed]
- Liu SJ, Dang HX, Lim DA, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer 2021;21:446-60. [Crossref] [PubMed]
- Zhao Y, Yuan D, Zhu D, et al. LncRNA-MSC-AS1 inhibits the ovarian cancer progression by targeting miR-425-5p. J Ovarian Res 2021;14:109. [Crossref] [PubMed]
- Wang HM, Shen SL, Li NM, et al. LncRNA CDKN2BAS aggravates the progression of ovarian cancer by positively interacting with GAS6. Eur Rev Med Pharmacol Sci 2020;24:5946-52. [PubMed]
- Pei C, Gong X, Zhang Y. LncRNA MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via sponging microrna-22. Am J Transl Res 2020;12:6977-87. [PubMed]
- Zheng ZJ, Liu Y, Wang HJ, et al. LncRNA SNHG17 promotes proliferation and invasion of ovarian cancer cells through up-regulating FOXA1. Eur Rev Med Pharmacol Sci 2020;24:9282-9. [PubMed]
- Wang X, Yang B, She Y, et al. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J Cell Biochem 2018;119:7790-9. [Crossref] [PubMed]
- Shen W, Xie X, Liu M, et al. Diagnostic Value of lncRNA ROR in Differentiating Ovarian Cancer Patients. Clin Lab 2020;66. [Crossref] [PubMed]
- Chen M, Lei N, Tian W, et al. Recent advances of non-coding RNAs in ovarian cancer prognosis and therapeutics. Ther Adv Med Oncol 2022;14:17588359221118010. [Crossref] [PubMed]
- Braga EA, Fridman MV, Moscovtsev AA, et al. LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms. Int J Mol Sci 2020;21:8855. [Crossref] [PubMed]
- Qu J, Kamal MA, Yuan C. The Regulatory Roles of Long Non-Coding RNA in the Chemoresistance Process of Ovarian cancer. Curr Pharm Des 2019;25:856-61. [Crossref] [PubMed]
- Wu Y, Gu W, Han X, et al. LncRNA PVT1 promotes the progression of ovarian cancer by activating TGF-β pathway via miR-148a-3p/AGO1 axis. J Cell Mol Med 2021;25:8229-43. [Crossref] [PubMed]
- Xu Q, Lin YB, Li L, et al. LncRNA TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer through enhancing TLR8 mRNA stability. Biochem Biophys Res Commun 2020;526:857-64. [Crossref] [PubMed]


