
Role of methylation-related genes CRYAB and SLC39A11 in the occurrence and development of lung adenocarcinoma
Introduction
Human crystallin alpha B (CRYAB), also known as human alpha B-crystallin (HspB5) or αB-crystallin, is a subtype of small heat shock protein (sHsp). It was first discovered in the lens of the eye and has been linked to a variety of diseases, including eye disease, heart disease and even breast, lung, prostate and ovarian cancers (1). CRYAB comprises of an N-terminal domain, a central domain, and a C-terminal domain (2). As a chaperone, CRYAB plays an important role in inhibiting the aggregation of denatured proteins and maintaining the pool of non-natural foldable intermediates of easily aggregable proteins in large soluble polymeric structures (3). Ectopic expression of CRYAB in various cell types may provide protection against apoptotic stimuli, oxidative stress, and anticancer drug exposure (4,5). Meanwhile, RNA interference can silence its expression and consequently sensitize cells to apoptosis. Previous research has demonstrated the high expression of CRYAB in human cancers and the significant association between CRYAB and poor prognosis of cancer patients (6). Apoptosis is a programmed death that is negatively regulated by sHsps (7,8). In contrast to the increased expression of sHsps in response to environmental damage such as heat shock, the expression of these proteins was not upregulated in apoptosis. In some cells, apoptosis mediated by immune system or therapeutic drugs can be suppressed by expressed sHsps (9). CRYAB is currently supposed to interact with and regulate the activity of specific proteins in the initiation and execution stages of apoptosis. CRYAB is an antiapoptotic protein whose main property is to negatively regulate Bax and caspase-3, 2 proapoptotic members of the B-cell lymphoma 2 (Bcl-2) family (10). CRYAB interplays directly with caspase-3, Bax, and Bcl-xS. Furthermore, it can block their transformation from cytoplasm to mitochondria and consequently ward off stress-induced apoptosis (11). Analogously, CRYAB interplays with p53 to seclude its translocation to the mitochondria, thus indirectly suppressing its proapoptotic effect on the apoptotic Bcl-2 molecule (12). CRYAB prevents apoptosis by inhibiting the activation of caspase-3 and poly (ADP-ribose) polymerase (PARP). Furthermore, the incubation of immortalized astrocytes or microglia cell lines with CRYAB significantly inhibited inflammatory responses mediated by astrocytes and microglia in either autocrine or paracrine manner. A previous study has demonstrated that high expression of CRYAB was correlated with poor survival and TNM stage in NSCLC patients. CRYAB expression may serve as a novel prognostic marker in NSCLC patients (13).
Solute carriers (SLCs) are important transmembrane transporters, with members organized into 65 families. SLC participates in a series of physiological processes and may contribute to the identification of new therapeutic targets for human malignant tumors (13,14). The SLC39A family of zinc transporters play a key role in maintaining intracellular zinc homeostasis. The intracellular zinc transporters includes 2 major families: the Zn transporter (ZnT) family (SLC30A) and the Zrt- and Irt-like protein (ZIP) family (SLC39A) (15). SLC39A helps increase cytosolic zinc levels by transporting zinc from the extracellular environment or intracellular pools, while the ZnT transporter facilitates zinc transport in the opposite direction (16). There are 4 subfamilies of the ZIP transporters [i.e. subfamily I (ZIP9), subfamily II (ZIP1–3), LIV-1 subfamily (ZIP4–8, 10, and 12–14), and gufA (ZIP11)] (17). It has been reported that the SLC39 gene plays a key role in colorectal cancer (CRC), esophageal cancer, prostate cancer (Pca), lung cancer, hepatocellular carcinoma, and other malignant tumors (18,19). In addition, studies have shown that certain parts of the SLC39 gene may be associated with survival in patients with certain types of cancer and may be used as a biomarker for cancer diagnosis or prognosis. For example, by analyzing specimens obtained by surgery or endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA), Xu et al. found that SLC39A4 could be used as a prognostic and diagnostic marker for pancreatic cancer (20). Ding et al. (21) assessed the prognostic value of all SLCA39 genes for gastric cancer (GC). It was proved that SLC39A7 played an essential role for cell survival of lung adenocarcinoma (LUAD). SLC39A groups were significantly associated with prognosis of LUAD. The SLC39A7 gene was significantly linked with survival and growth of LUAD cells (22). However, few studies have explored the expression and prognostic value of the SLC39A11 gene in LUAD.
DNA methylation regulates and controls gene expression by recruiting proteins associated with gene repression or by blocking the binding between transcription factor(s) and DNA (23). CRYAB and SLC39A11 genes are supposed to related to DNA methylation. The abnormal epigenetic regulation in cancer comprises DNA methylation, miRNA gene silencing, histone methylation, mRNA and non-coding RNA methylation, and histone acetylation (24). DNA methylation is closely related to the prognosis of LUDA. Although a study has proved aberrant DNA methylation levels in LUAD, a comprehensive regulatory network and pathway analysis of DNA methylation levels and miRNA epigenetic alterations has not been performed (25). The 2 genes investigated in the current study, SLC39A11 and CRYAB, were silenced by small interfering RNA (siRNA). The efficiency of siRNA interference was then detected using reverse transcription-polymerase chain reaction (RT-PCR). Next, Cell Counting Kit-8 (CCK-8) assay, Transwell migration assay, flow cytometry, and clone proliferation assay were performed to detect the proliferation, migration, and cell cycle of lung adenocarcinoma tumor cell lines A549 and H1975 after interference with CRYAB and SLC39A11 genes in order to determine whether the genes were related to the proliferation of lung adenocarcinoma and whether they could be used as potential therapeutic targets and/or for prognosis prediction in patients with lung adenocarcinoma. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3576/rc).
Methods
Materials
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). CRYAB and SLC39A11 genes, which are related to prognosis and methylation, were identified from The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) database. siRNA was purchased from Shanghai GenePharma Co., Ltd., Shanghai, China. We constructed the siRNA-CRYAB-A549, siRNA-CRYAB-H1975, siRNA-SLC39A11-A549, and siRNA-SLC39A11-H1975 cell lines ourselves.
Experimental methods
Cell culture
The temperature of the water bath was set to 37 °C in advance, and 1640 medium (Hyclone, Logan, UT, USA) and serum were thawed or rewarmed in the 37-degree water bath for 30 minutes. The A549 and H1975 cells were retrieved from a liquid nitrogen tank or −80 °C refrigerator. After the screw tops of the cryovials were tightened, the cryovials were thawed by rolling them rapidly in the water bath until the cells were completely thawed into a liquid state. The cryovials were then wiped dry before being placed in a pass box. In the cell culture room, 50 mL of complete medium supplemented with 10% fetal bovine serum (FBS; Zhejiang Tianhang Biological Technology Co., Ltd., Zhejiang, China) was prepared. Next, 45 mL of 1640 medium was transferred into a 50-mL centrifuge tube via a 50-mL pipette using an electric pipette, and 5 mL of serum FBS was transferred to a centrifuge tube via a 5-mL pipette. The media were gently mixed, and the complete medium was labeled with the date. The cells in the cryovials were pipetted into a 15-mL centrifuge tube, in which 1 mL of complete medium was added before rinsing and cryopreservation. The rinsed mixture was transferred to a 15-mL centrifuge tube, washed twice, and then centrifuged at 800 rpm for 5 minutes at 25 °C. Complete medium was added after the supernatants were removed. The cells were gently resuspended using a 1-mL pipette. After the cell concentration was calculated, the cell suspension was transferred to a 25-cm2 cell culture flask (Thermo Fisher Scientific, Waltham, MA, USA) to culture the cells. The volume of the culture medium was increased to 5 mL again, and the flask was placed in an incubator at 37 °C and 5% CO2.
Cell transfection experiment
The morphology and density of the cells were observed under a microscope. When the morphology was normal and the density reached 40–50%, the siRNA interference experiments were carried out. The experiment was performed in a 6-well plate, and 7.5 µL Lipofectamine 3000 (Lipo3000) was mixed with 125 µL Opti-MEM and left to stand at room temperature for 5 minutes. siRNA (5 µg) was added to 250 µL Opti-MEM, mixed well with a pipette, and left to stand at room temperature for 5 minutes. These 2 mixtures were mixed with a pipette at a ratio of 1:1 and then incubated at room temperature (25 °C) for 10–15 minutes. The mixture was placed into a culture dish, and subsequently the cells were cultured in a cell incubator (Thermo Electron, San Jose, CA, USA) at 37 °C and 5% CO2.
RNA extraction
Cells were collected from the 37 °C incubator, and the medium was gently removed with a pipette. Next, 1 mL of cell-specific phosphate-buffered saline (PBS; Sigma-Aldrich, St Louis, MO, USA) was added, and the cells were washed 3 times. At the last washing, the PBS was removed with a pipette. TRIzol (500 µL per well) was added to a 6-well plate and left to stand on ice for 5 minutes. The lysed cells were transferred to a 1.5-mL Eppendorf (EP) tube. Chloroform (200 µL; Invitrogen, Carlsbad, CA, USA) was added to the EP tube, mixed gently, incubated at room temperature for 5 minutes, and centrifuged at 12,000 rpm at 4 °C for 15 minutes. The supernatant was transferred to a fresh EP tube, in which 500 µL isopropanol was added and mixed well before being placed on ice for 10 minutes. After the mixture was centrifuged again at 12,000 rpm at 4 °C for 10 minutes, the supernatant was removed and 1 mL of 75% ethanol was added. After the precipitate was washed, the mixture was centrifuged again at 7,500 rpm at 4 °C for 10 minutes and then air dried. Finally, 20–50 µL of diethylpyrocarbonate (DEPC)-treated water was added, mixed well, and the concentration was measured with NanoDrop.
Reverse transcription
Reverse transcription and RT-PCR were performed using a reverse transcription kit (TAKARA STANDARD Co., Ltd., Osaka, Japan) in strict accordance with the manufacturer’s instructions. The first step was to remove the genomic DNA. The corresponding volumes were calculated in turn, and the corresponding reagents were added into 100-µL small EP microtubes. Subsequently, the reagents were mixed well by vortexing, and all liquids were centrifuged. The PCR program was set at 42 °C for 2 minutes. The RNA from which genomic DNA had been removed was reverse transcribed. According to the following 20-µL system, the corresponding volume of reagents were added, and the reverse transcription program was set at 37 °C for 15 minutes, 85 °C for 5 seconds, and finally 4 °C.
RT-PCR
Real-time fluorescence quantitative PCR was performed according to the standard procedure of the TAKARA kit. The concentration of the complementary DNA (cDNA) generated after reverse transcription was determined. Subsequently, the sample was loaded according to the system. The first step of the program was set at 95 °C for 30 seconds, the second step was 40 cycles of 95 °C for 5 seconds and 60 °C for 31 seconds, and the last step was 95 °C for 15 seconds, 60 °C for 1 minute, and 95 °C for 15 seconds. RT-PCR results were calculated using the −ΔΔCT method. The primer sequences used in this study are shown in Table S1.
CCK-8 assay
CCC-8 reagent was purchased from Nanjing KeyGEN Biotech Co., Ltd. (Nanjing, China). Under the action of 1-methoxy-5-methylphenazinium methyl sulfate (1-methoxy PMS), its main component 2-(2-methoxyl)-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfobenzene)-2H-tetrazolium monosodium is reduced to highly water-soluble orange-yellow formazan by dehydrogenase in mitochondria. The concentration of the formazan complex can be read at an optical density (OD) value of 450 nm, which can be used to detect cell viability, proliferation, or toxicity. At the beginning of our experiment, after siRNA was added for interference, the cells were trypsinized, resuspended, re-added to a 96-well plate (100 µL per well), and cultured at 37 °C in an incubator supplied with 5% CO2. Next, 10 µL of CCK-8 reagent was added 1, 2, 3, 4, 5, and 6 days after culturing, and the cells were incubated in the dark at 37 °C for 1–4 hours. The mixture was mixed well on the shaker, and then the absorbance was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell cell invasion assay
Cell invasion and migration assays were done using the Corning’s Transwell® system (KeyGEN Biotech) according to the manufacturer’s instructions. Two lung adenocarcinoma tumor cell lines were transfected with siRNA and cultured in the incubator for 24 hours. The cells were first digested and centrifuged using the cell passage method or cell cryopreservation method. After the supernatant was removed, the cells were resuspended in serum-free Roswell Park Memorial Institute (RPMI) 1640 medium and pipetted evenly with a 1-mL pipette. The total cells were counted with a cell counter. After 1×104 cells were added into each Transwell chamber, the volume of resuspended cell solution was calculated and the corresponding volume of cells was added onto the upper insert of the chamber. If the volume was below 200 µL, it was supplemented with serum-free RPMI 1640 medium to reach 200 µL. Next, 700 µL of complete medium (supplemented with 10% of serum-free RPMI 1640) was added to the lower insert of the chamber. This step was completed in a gentle manner to avoid the appearance of air bubbles. The chamber was placed in a cell incubator with a constant temperature of 37 °C for 24 hours. The upper insert of the Transwell chamber was taken out with clean tweezers that had been autoclaved, and the cells were washed several times with PBS. The cells were fixed with 4% paraformaldehyde, placed at room temperature for 1 hour, and rinsed with PBS again after fixation. The liquid in the upper insert and cells that had not migrated were removed as much as possible using cotton swabs. The cells were then stained with crystal violet for 10 minutes, after which they were rinsed again with PBS. The insert was wiped clean and transferred to the microscope. The results were photographed, and more than 3 visual fields were randomly selected for further statistical analysis.
Apoptosis assay
Under normal physiological conditions, phosphatidylserine (PS) is distributed exclusively on the inner side of the cell membrane’s lipid bilayer. In contrast, during the early stages of apoptosis, PS flip-flops out. The exposure of PS on the outer plasma membrane is not a unique feature of apoptotic cells. Rather, it can also occur in necrotic cells. Nevertheless, the cell membrane is intact in the early stage of apoptosis, while the cell membrane integrity has been destroyed in the early stage of cell necrosis. Annexin V is a Ca2+-dependent phospholipid-binding protein that has a high affinity for PS and therefore can be used as a sensitive marker of early apoptosis. The annexin V was labeled with fluorescein isothiocyanate (FITC) and then used as a fluorescent probe to detect the occurrence of apoptosis by flow cytometry. Propidium iodide (PI) is a nucleic acid dye that is unable to penetrate intact cell membrane. However, for cells in the middle or late stages of apoptosis and necrotic cells, it can penetrate the cell membrane and stain the nucleus. Therefore, double-staining with annexin V and PI can distinguish cells in different apoptotic stages and detect the integrity of the cell membrane. The specific detection method was as follows:
Cell treatment
siRNA interference experiments were performed on lung adenocarcinoma cell lines. After having been cultured in an incubator for 24 hours, the cells were digested with ethylene diamine tetraacetic acid (EDTA)-free trypsin for a relatively short period of time. After digestion was complete, the mixture was centrifuged at 1,500 rpm for 5 minutes at room temperature. The supernatant was discarded, and the cells were resuspended in PBS and then centrifuged again under the same conditions. The cells were washed twice, and the cell precipitate was collected.
Preparation of 1 × Annexin V working solution
The 5 × Annexin V-FITC binding buffer was diluted with filter-sterilized deionized water to 1 × Annexin V working solution.
Addition of 100 µL of 1 × V-FITC binding buffer to resuspend the cells
After 5 µL of 1 × Annexin V staining solution and 5 µL of 100 µg/mL PI staining solution were added, the cells were incubated in the dark at room temperature for about 15 minutes.
The mixture was supplemented with 400 µL of V-FITC binding buffer and gently pipetted to mix well. The single-cell suspension was filtered through a 300-mesh sieve and detection was performed on a flow cytometer. The data were analyzed using BD FACS software.
Plate clone formation assay
A549 and H1975 cells in logarithmic growth phase were taken from each group, digested with 0.25% trypsin for a specific period of time, and resuspended to make a single-cell suspension. After the volume was calculated according to the number of cells, the cells were inoculated into 6-well plates at a density of 500 cells/cell. The plates were placed in a 37 °C, 5% CO2 incubator for a specific period of time and observed regularly until clonal cells visible to the naked eye appeared. The plates were removed and washed with PBS. The cells were fixed in 20% methanol at room temperature for 12 minutes, and then the methanol was removed. After washing with PBS, the cells were stained with 2.5% crystal violet solution for 15 minutes. After staining, the cells were washed and air dried. The dried cells were observed under a microscope, and the clone formation rate was calculated according to the number of observed cell clones. The clone formation rate (%) = (the number of clones divided by the number of inoculated cells) × 100%.
Cell cycle analysis
The cell cycle changes in each treatment group were detected by flow cytometry. The cell cycle is the entire process that continuously dividing cells go through from the end of one mitosis to the beginning of the next mitosis. After the cell has completed this process, the genetic materials inside the original cell are duplicated and doubled, and the genetic materials are distributed equally between the 2 daughter cells during cell division. A cell cycle consists of mitosis (M) and interphase [gap 1 (G1), synthesis (S), and gap 2 (G2) phases]. The entire cell cycle progression can be expressed as G1→S→G2→M. Detection of DNA cycle changes can be used to reflect the cell cycle, that is, the state of cell proliferation. One DNA-binding dye is PI, which is a fluorescent dye. Since the DNA content of cells differs in different stages, when a fluorescent dye is used, the amount of fluorescent dye bound to cells in different stages is also different, and accordingly, the fluorescence intensity detected by the cytometer also differs. Therefore, observing changes in cell fluorescence intensity during flow cytometry can characterize changes in the cell cycle. Corresponding siRNA was transfected into A549 and H1975 cell lines. After having been cultured in a constant-temperature incubator for 24 hours, the cells were harvested, washed with PBS, and digested with trypsin. Subsequently, the cells were collected, washed with PBS, and counted. After the cell concentration was adjusted to 1×106/mL, 1 mL of each sample was collected for centrifuging. The supernatant was then discarded, 1 mL of 70% cold ethanol was added and mixed well, and the cells were fixed at 4 °C for more than 3 hours. PBS (2 mL) was added, the mixture centrifuged, and the supernatant discarded. The cells were supplemented with 400 µL of PI solution, mixed well, and stored at room temperature in the dark for 30 minutes. Finally, data were collected on a flow cytometer. Analysis was performed using Mod Fit LT software (Verity Software House Inc., Topsham, ME, USA), and 10,000 cells were analyzed per sample. Single-cell populations were selected using an appropriate gating strategy and applied for all samples. Measurements were made on standardized cell populations.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software. The measurement data are expressed as mean ± standard deviation (). The data of 2 groups were compared by t-test, and comparisons among 3 or more groups were based on one-way analysis of variance (ANOVA). A P value of less than 0.05 was considered to be statistically significant.
Results
Differentially-expressed genes (DEGs) filtered from TCGA and GEO databases
Based on TCGA lung adenocarcinoma data, the gene of CRYAB and SLC39A11 were downregulated and upregulated in lung adenocarcinoma tissues and the differences were significantly (Figure 1A). Accordingly, the same phenomenon occured in GSE32863 (Figure 1A).
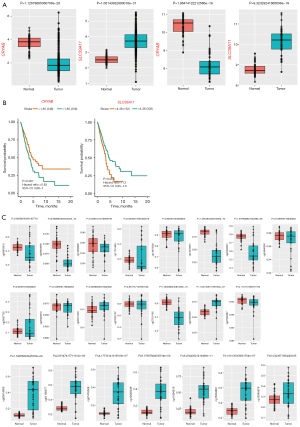
What’s more, based on TCGA data of patient survival and gene expression, the potential correlations between 2 DEGs and survival of lung adenocarcinoma patients were explored. The patients with high SLC39A11 expression (>4.38) had a significantly lower survival rate than those with low SLC39A11 expression (<4.38) (P=0.003), and the survival rate of patients with CRYAB expression less than 1.85 was lower than that of patients with CRYAB expression greater than 1.85 (P=0.007) (Figure 1B). The data also showed that the corresponding methylation sites of the two genes were also dysregulated, and there was a statistical difference (P<0.05) (Figure 1C).
Validation of siRNA interference efficiency of CRYAB and SLC39A11
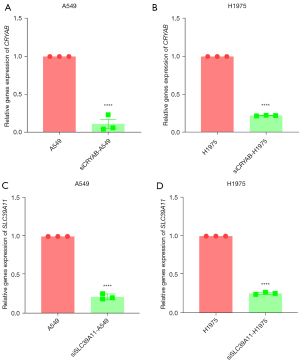
Lung adenocarcinoma cell lines A549 and H1975 were subjected to siRNA interference and were then cultured for 24 hours. Subsequently, RNA was extracted from each group for reverse transcription and RT-PCR. The results showed that compared with the untreated A549 cell control group, the siCRYAB-transfected A549 and H1975 cell lines had decreased messenger RNA (mRNA) expression levels of CRYAB (Figure 2A,2B). Compared with the untreated A549 and H1975 controls, the mRNA expression level of SLC39A11 was decreased in both the siSLC39A11-transfected A549 cell line and the siSLC39A11-transfected H1975 cell line (Figure 2C,2D). Therefore, siRNA successfully interfered with the mRNA expression of SLC39A11 and CRYAB in cell lines A549 and H1975. The transfection assay was successful, and the products could be used in subsequent experiments.
Effects of CRYAB and SLC39A11 knockout on the proliferation of lung adenocarcinoma cells
The CCK-8 method was used to detect the effect of knockout of CRYAB and SLC39A11 on cell proliferation in A549 and H1975 cell lines. Cells were collected 1, 2, 3, 4, 5, and 6 days after knockdown of CRYAB and SLC39A11, respectively. The CCK-8 staining solution was added dropwise, and then the cells were observed under a microscope. In the A549 lung adenocarcinoma cell line, the mean OD values 1–6 days after CRYAB knockout were higher than those of the control group at 1–6 days (Figure 3A). In the H1975 lung adenocarcinoma cell line, the mean OD values 1–6 days after CRYAB knockout were higher than those of the control group at 1–6 days (Figure 3B). Therefore, knocking out CRYAB in lung adenocarcinoma cell lines A549 and H1975 promoted the proliferation of lung adenocarcinoma cells. The average OD values of A549 cells after SLC39A11 knockout were lower than those of the control group. In the H1975 cell line, the mean OD values 1–6 days after SLC39A11 knockout were lower than those of the control group (Figure 4). Thus, knockdown of SLC39A11 inhibited the proliferation of lung adenocarcinoma cells in A549 and H1975.


Effects of CRYAB and SLC39A11 knockout on the formation of lung adenocarcinoma clones
After the knockout of CRYAB and SLC39A11 in lung adenocarcinoma cell lines A549 and H1975, the cells were placed in an incubator for a period of time. When the lung adenocarcinoma cells were in the logarithmic growth phase, the cells were collected for plate clone formation assay, during which the changes in the clone growth of lung adenocarcinoma cell lines were observed. In the A549 cell line, the mean number of clones of lung adenocarcinoma cells after knocking out CRYAB was 389, which was significantly higher than that (n=292) of the control group (P<0.05) (Figure 5A-5C). Thus, knockout of CRYAB promoted the cloning of lung adenocarcinoma cell line A549. In addition, the mean number of clones of lung adenocarcinoma cells in the A549 cell line after knocking out SLC39A11 was 362, which was significantly lower than that (n=499) of the control group (P<0.05) (Figure 5A-5C). Therefore, knockdown of SLC39A11 inhibited the cloning of lung adenocarcinoma cell line A549.
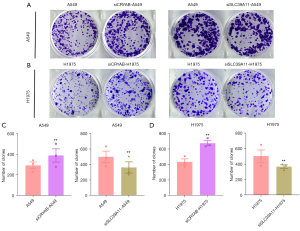
Similar effects were observed in the lung adenocarcinoma cell line H1975. In the H1975 cell line, the mean number of plate clones of H1975 after knocking out CRYAB was 675, which was significantly higher than that (n=434) of the control group (P<0.05). The number of clones in H1975 cells after knocking out SLC39A11 was 370, which was lower than that (n=505) in the control group (Figure 5A,5B,5D). Therefore, knockdown of CRYAB in H1975 cell line also promoted the cloning of lung adenocarcinoma cells, and knockdown of SLC39A11 also suppressed the cloning of lung adenocarcinoma cells.
Effects of CRYAB and SLC39A11 knockout on apoptosis of lung adenocarcinoma cells
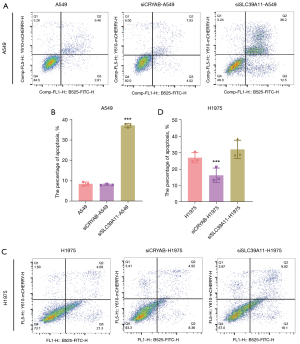
The results of plate cloning assay showed that knockout of CRYAB and SLC39A11 had significant effects on the cloning of lung adenocarcinoma cells. To further observe the effects of siCRYAB and si-SLC39A11 on the apoptosis of lung adenocarcinoma cells, apoptosis detection kit combined with flow cytometry was used. In the A549 cell line, after CRYAB and SLC39A11 were knocked out, the average proportion of lung adenocarcinoma cells in the apoptotic phase was 8.16% and 37.20% respectively (Figure 6A), and the proportion of cells in the control group was 8.53% (Figure 6B). In the H1975 cell line, after CRYAB and SLC39A11 were knocked out, the average proportion of lung adenocarcinoma cells in the apoptotic phase was 16.20% and 31.98% (Figure 6C), respectively, and the proportion of cells in the control group was 26.92% (Figure 6D). Therefore, in both A549 and H1975 cell lines, knockout of CRYAB inhibited the apoptosis of lung adenocarcinoma cells, whereas knockout of SLC39A11 promoted the apoptosis of lung adenocarcinoma cells.
Effects of CRYAB and SLC39A11 knockout on the cell cycle of lung adenocarcinoma cells
The CRYAB and SLC39A11 genes were knocked out in lung adenocarcinoma cell lines A549 and H1975, and the results were observed by apoptosis detection kit Annexin V-PE combined with flow cytometry. In the A549 cell line, after CRYAB was knocked out, flow cytometry revealed that the proportion of cells in G0/G1 phase was 49.15%, the proportion of cells in S phase was 45.57%, and the proportion of cells in G2/M phase was 5.28%. The proportions of cells in G0/G1, S, and G2/M phases after SLC39A11 knockout were 56.85%, 37.41%, and 5.74%, respectively. In contrast, the proportions of cells in G0/G1, S, and G2/M phases were 60.14%, 32.67%, and 7.19%, respectively, in the control group. Compared with the control group, the number of cells arrested in G1-G2 after CRYAB knockout was significantly reduced, indicating that knockout of CRYAB promoted A549 tumor cells to enter the cell division phase, whereas knockout of SLC39A11 increased the number of cells in interphase and decreased the number of cells in the division phase; that is, it slowed down the tumor cell division cycle (Figure 7A).
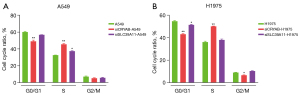
In H1975 cells, the proportions of cells in G0/G1, S, and G2/M phases were 43.19%, 50.17%, and 6.65%, respectively, after CRYAB knockout, and the proportions of cells in G0/G1, S, and G2/M phases were 51.49%, 38.08%, and 10.43%, respectively, after SLC39A11 knockout. In contrast, the proportions of cells in the G0/G1, S, and G2/M phases were 54.78%, 36.19%, and 9.04%, respectively, in the controls (Figure 7B). Thus, in the H1975 cell line, knockout of CRYAB also lowered the proportion of cells in interphase and increased the proportion of mitotic cells, while knockout of SLC39A11 also slowed down the division cycle of tumor cells.
Effects of knockout of CRYAB and SLC39A11 on invasion and metastasis of lung adenocarcinoma cells
In order to explore the roles of CRYAB and SLC39A11 in the invasion and metastasis of lung adenocarcinoma cells, we observed the invasion and metastasis of lung adenocarcinoma cells in a Transwell chamber after knocking out CRYAB and SLC39A11 in tumor cell lines A549 and H1975.
In the A549 cell line, the number of migrating cells with CRYAB knockout in the Transwell chamber was significantly higher than that of untreated A549 cells in the Transwell chamber (Figure 8). In the H1975 cell line, similar results were observed, with the number of siCRYAB-interfered migrating cells in the Transwell chamber significantly larger than that of the controls (Figure 8). Therefore, in both A549 and H1975 cell lines, lung adenocarcinoma cells with CRYAB knockout had stronger migration ability than controls; in other words, knockdown of CRYAB promoted the migration of lung adenocarcinoma cells.
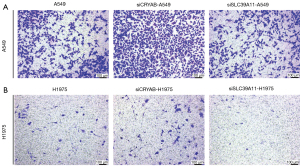
In the A549 cell line, the number of migrating cells after SLC39A11 knockout in the top insert of the Transwell chamber was significantly higher than that of the untreated A549 cells (Figure 8). Thus, knockdown of SLC39A11 inhibited the migration of lung adenocarcinoma cells in A549 cell line. In the H1975 cell line, similar results were observed, with the number of siCRYAB-interfered cells in the Transwell chamber significantly smaller than that of the untreated cells (Figure 8). Therefore, in the H1975 cell line, knockout of SLC39A11 also reduced the invasive ability of lung adenocarcinoma cells.
Discussion
The incidence of lung adenocarcinoma, the most common subtype of non-small cell lung cancer, is still rising. In the earliest stage of lung adenocarcinoma, the tumor is characterized by ground glass nodules on computed tomography (CT). These preinvasive nodules can develop into invasive lung adenocarcinoma over time. However, due to the low efficiency of CT in screening lung adenocarcinoma, most lung adenocarcinomas are detected at an advanced stage, leading to a high mortality rate and significant treatment expenditure. The newly discovered genomic features of cancers over the past few decades have provided early detection methods, refined risk assessments, and led to therapeutic interventions for lung cancer. However, the role of these strategies is limited. The poor understanding of the biological characteristics of lung adenocarcinoma results in poor prognosis and low survival rates in most lung adenocarcinoma patients (26). In this study, we identified several candidate molecules and pathways related to the prognosis of lung adenocarcinoma, which could be employed as potential therapeutic targets.
Therefore, based on the TCGA data and survival results, we selected CRYAB and SLC39A11 for in vitro phenotypic and functional characterization to validate the values of these 2 genes in lung adenocarcinoma cell lines, aiming to find early diagnostic markers for lung adenocarcinoma and develop new therapeutic targets. First, data from TCGA revealed that CRYAB was significantly downregulated and SLC39A11 was significantly upregulated in lung adenocarcinoma tissues. The results of survival analysis indicated that the survival rate of lung adenocarcinoma patients with low CRYAB expression was lower than that of patients with high CRYAB expression, whereas SLC39A11 expression was negatively correlated with the survival rate. The proliferation, clone formation, cell cycle, apoptosis, and invasion of lung adenocarcinoma cells were detected after the knockout of CRYAB and SLC39A11 genes in lung adenocarcinoma cell lines A549 and H1975. CCK-8 assay revealed that CRYAB knockout promoted the proliferation of lung adenocarcinoma cell lines A549 and H1975, while SLC39A11 knockout inhibited the proliferation of lung adenocarcinoma cells. Plate cloning assay showed that CRYAB knockdown promoted the cloning of lung adenocarcinoma cells A549 and H1975, and SLC39A11 knockdown suppressed the cloning of lung adenocarcinoma cells. Further experiments of apoptosis and cell cycle changes showed that knockout of CRYAB in A549 and H1975 cells inhibited the apoptosis of lung adenocarcinoma cells and promoted lung adenocarcinoma cells to enter the division phase, while silencing of SLC39A11 promoted the apoptosis of lung adenocarcinoma cells and suppressed the cell cycle of lung adenocarcinoma cells. Finally, Transwell invasion assay showed that migratory ability was increased in lung adenocarcinoma cells with CRYAB knockout and decreased in lung adenocarcinoma cells with SLC39A11 knockout. Therefore, these assays demonstrated that CRYAB and SLC39A11 were related to the occurrence and development of lung adenocarcinoma, and the prognosis of lung adenocarcinoma patients could be predicted by monitoring the levels of CRYAB and SLC39A11.
Alpha-crystallin B is a chaperone member of the sHsp family. To date, it has been reported to protect cells from adverse conditions by regulating several survival and stress recovery-related cellular processes such as protein degradation, cytoskeleton stabilization, and apoptosis (27-29). Given the role of CRYAB in maintaining the physiological function of cells and organs, CRYAB can play a decisive role in the development of several pathological processes, including deproteinization-related myopathies (e.g., dilated and restrictive cardiomyopathy), cataract formation, neurodegenerative diseases, and cancers (30,31). In 2007, Liu et al. reported that CRYAB played an important role in lung adenocarcinoma (32). So far, CRYAB has been reported to interact with several tumor-associated proteins, such as p53, procaspase-3, Bax, fibroblast growth factor-2 (FGF2), and F-actin, which play key roles in tumor cell proliferation and the tumor cell cycle (33). The inclusion of CRYAB as a marker for identifying primary cancers was suggested in the early 1990s, although some preliminary studies did not find a definite association between the CRYAB expression in cancer cells and morphological changes (34,35). Most of the known properties (e.g., cytoprotective, antiapoptotic, proangiogenic, and prometastatic activities) of sHsp family members were also found in the current study.
CRYAB has also been found to mediate the migration and invasion of GC cells through nuclear factor (NF)-κB signaling pathway-mediated epithelial-mesenchymal transition (EMT), which may provide a new target for GC therapy (36). In 2020, a study identified 13 miRNAs associated with PCa metastasis through bioinformatics analysis. The authors also found through mechanistic studies that miR-671-5p directly bound to the 3’ untranslated regions of NFIA mRNA, and NFIA directly bound to the CRYAB promoter. High expression of NFIA and CRYAB negatively correlated with the advanced clinicopathological characteristics and metastasis status of PCa patients. Thus, miR-671-5p promoted PCa development and metastasis by suppressing the NFIA/CRYAB axis (37). In 2021, Deng et al. (2) used Kaplan-Meier survival curves from TCGA to evaluate the relationship between CRYAB expression and both overall survival and progression-free survival. The relationships between CRYAB expression and infiltrating immune cells and their corresponding gene marker sets were examined using the Tumor Immune Estimation Resource (TIMER) database. The expression of CRYAB in CRC tissues was significantly lower than that in normal tissues (P<0.05). High CRYAB gene expression and CRYAB gene methylation levels were associated with highly malignant or more advanced tumors, lymph node metastases, and (TNM) cancer staging.
Furthermore, in CRC, CRYAB expression was positively associated with immune infiltrating cells, such as neutrophils, macrophages, cluster of differentiation (CD)8+ T cells, and CD4+ T cells, as well as immune-related genes, including CD2, CD3D, and CD3E. The methylation sites CG13084335, CG15545878, CG13210534, CG15318568 were negatively correlated with CRYAB expression (2). A recent clinical trial investigated the inhibitory effect of sHsps when used as molecular targets for tumor therapy (38). However, the heat shock proteins are a ubiquitous class of ATP-independent chaperones, which makes the targeting of these small molecules particularly difficult (39). Therefore, further basic and clinical studies are required concerning CRYAB-targeted therapy for lung adenocarcinoma.
SLC39A11, a protein-coding gene, was also investigated in the current study. SLC39A11 has been reported to be associated with Brown-Vialetto-Van Laere syndrome 2 and acrodermatitis (40). The relevant pathways include the metal ion SLC transporters and the nuclear factor erythroid 2–related factor 2 (Nrf2) pathway, and its gene ontology (GO) annotations include metal ion transmembrane transporter activity. The protein encoded by SLC39A11 can be used as a component of cell membrane, and as a key protein for transporting metal ion zinc, it plays an important role in maintaining the balance of zinc ion inside and outside the cells (41). As seen in the current study, SLC39A11 also plays a role in promoting tumor cell proliferation and migration in the occurrence and development of other cancers. In 2015, Wu et al. (42) demonstrated the correlationships of relevant proteins with the prognosis of patients with bladder cancer. In 2015, a multicenter study of ovarian cancer obtained DNA samples from 14,525 patients with invasive epithelial ovarian cancer (EOC) and 23,447 controls. A total of 279 single nucleotide polymorphisms (SNPs), representing 131 genes, were genotyped using an Illumina Infinium iSelect BeadChip as part of the Collaborative Oncological Gene-environment Study (COGS). Under a log-additive model, SNP analyses were performed by using unconditional logistic regression, and the false discovery rate (FDR) q<0.2 was employed for adjusting multiple comparisons. The results showed that SLC39A11 was associated with the malignant potential of ovarian cancer (43). In 2018, Barresi et al. (44) revealed altered zinc transporter expression profiles in CRC by transcriptome analysis. The results showed that specific ZnT and ZIP transcripts were up-regulated in CRC. The upregulated SLC30A5, SLC30A6, and SLC30A7 transcripts, encoding zinc efflux transporters ZnT5, ZnT6, and ZnT7, which were localized on endoplasmic reticulum membranes, might be a portion of the coordinated transcriptional program associated with the increased early secretory pathway activity, while several transcriptional upregulated specific ZIP transporters, including SLC39A6, SLC39A7, SLC39A9, SLC39A10, and SLC39A11, may help to meet the increased demand of cancer cells for zinc. SLC30A9 is a nuclear receptor coactivator involved in the transcriptional regulation of Wnt-responsive genes. The exon-level analysis of SLC30A9 demonstrated the differential expression of alternative transcripts between CRC and normal colon mucosa. In 2019, Ding et al. (21) found mRNA deregulation was an important single factor for alteration in a variety of GC subtypes by using genetic alteration frequency analysis. Compared with normal gastric tissues, 14 out of 39 genes of the SLC family in the UALCAN database were significantly upregulated in GC tissue, and SLC39A4, SLC39A5, SLC39A6, and SLC39A10 mRNA expressions in the Oncomine database were higher as well. The survival analysis showed that the majority of the SLC family 39 genes were closely associated with the prognosis of GC patients with the SLC39A7, SLC39A11, and SLC39A14 correlated with favorable survival outcomes. The authors concluded that these 14 genes are potential prognostic markers for GC and may become new therapeutic targets (44). In 2020, by using the UALCAN database, Liu et al. evaluated the mRNA expressions of SLC family 39 genes in breast cancer (BC). The results of the study demonstrated that, compared with normal breast tissues, SLC39A1, SLC39A3, SLC39A4, SLC39A5, SLC39A6, SLC39A7, SLC39A9, SLC39A10, SLC39A11, and SLC39A13 were significantly upregulated in BC tissues. In subgroup analysis, most of the abnormally expressed SLC family members were correlated with prognoses of patients with specific types of BC. These results indicated that SLC family 39 members were promising biomarkers for BC prognosis (45). In the current study on lung adenocarcinoma, the expression of SLC39A11 was upregulated in lung adenocarcinoma tissue, which was consistent with the expression level of SLC39A11 in many other tumors. We concluded that SLC39A11 expression could also be used as a prognostic indicator and potential therapeutic target for lung adenocarcinoma.
Although the current study achieved similar findings to other tumor studies, it had some limitations. For example, we did not explore the mechanisms (e.g., specific molecules or signaling pathways) via which CRYAB and SLC39A11 exert their roles, which may limit the application of CRYAB and SLC39A11 in clinical research.
Conclusions
Knockout of CRYAB and SLC39A11 in lung adenocarcinoma cell lines A549 and H1975 exerted certain effects, indicating that CRYAB and SLC39A11 could be used as prognostic indicators and therapeutic targets for lung adenocarcinoma.
Acknowledgments
We would like to thank the National Natural Science Foundation of China for supporting this study.
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 81970167 and 81800108).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3576/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3576/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3576/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang K, Wu Y. A prognosis-related molecular subtype for early-stage non-small lung cell carcinoma by multi-omics integration analysis. BMC Cancer 2021;21:128. [Crossref] [PubMed]
- Deng J, Chen X, Zhan T, et al. CRYAB predicts clinical prognosis and is associated with immunocyte infiltration in colorectal cancer. PeerJ 2021;9:e12578. [Crossref] [PubMed]
- Zhang J, Liu J, Wu J, et al. Progression of the role of CRYAB in signaling pathways and cancers. Onco Targets Ther 2019;12:4129-39. [Crossref] [PubMed]
- Parcellier A, Schmitt E, Brunet M, et al. Small heat shock proteins HSP27 and alphaB-crystallin: cytoprotective and oncogenic functions. Antioxid Redox Signal 2005;7:404-13. [Crossref] [PubMed]
- Rajagopal P, Tse E, Borst AJ, et al. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. Elife 2015;4:e07304. [Crossref] [PubMed]
- Tao X, Cheng L, Li Y, et al. Expression of CRYAB with the angiogenesis and poor prognosis for human gastric cancer. Medicine (Baltimore) 2019;98:e17799. [Crossref] [PubMed]
- Singh MK, Sharma B, Tiwari PK. The small heat shock protein Hsp27: Present understanding and future prospects. J Therm Biol 2017;69:149-54. [Crossref] [PubMed]
- Obeng E. Apoptosis (programmed cell death) and its signals - A review. Braz J Biol 2021;81:1133-43. [Crossref] [PubMed]
- Cesa LC, Shao H, Srinivasan SR, et al. X-linked inhibitor of apoptosis protein (XIAP) is a client of heat shock protein 70 (Hsp70) and a biomarker of its inhibition. J Biol Chem 2018;293:2370-80. [Crossref] [PubMed]
- Malin D, Petrovic V, Strekalova E, et al. αB-crystallin: Portrait of a malignant chaperone as a cancer therapeutic target. Pharmacol Ther 2016;160:1-10. [Crossref] [PubMed]
- Hu WF, Gong L, Cao Z, et al. αA- and αB-crystallins interact with caspase-3 and Bax to guard mouse lens development. Curr Mol Med 2012;12:177-87. [Crossref] [PubMed]
- Tan L, Sha L, Hou N, et al. High α B-crystallin and p53 co-expression is associated with poor prognosis in ovarian cancer. Biosci Rep 2019;39:BSR20182407. [Crossref] [PubMed]
- Qin H, Ni Y, Tong J, et al. Elevated expression of CRYAB predicts unfavorable prognosis in non-small cell lung cancer. Med Oncol 2014;31:142. [Crossref] [PubMed]
- Lin L, Yee SW, Kim RB, et al. SLC transporters as therapeutic targets: emerging opportunities. Nat Rev Drug Discov 2015;14:543-60. [Crossref] [PubMed]
- Bai X, Moraes TF, Reithmeier RAF. Structural biology of solute carrier (SLC) membrane transport proteins. Mol Membr Biol 2017;34:1-32. [Crossref] [PubMed]
- Wang J, Zhao H, Xu Z, et al. Zinc dysregulation in cancers and its potential as a therapeutic target. Cancer Biol Med 2020;17:612-25. [Crossref] [PubMed]
- Kambe T, Tsuji T, Hashimoto A, et al. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol Rev 2015;95:749-84. [Crossref] [PubMed]
- Thomas P, Pang Y, Dong J, et al. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology 2014;155:4250-65. [Crossref] [PubMed]
- Wu DM, Liu T, Deng SH, et al. SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer. Sci Rep 2017;7:7211. [Crossref] [PubMed]
- Xu C, Wallace MB, Yang J, et al. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: a systemic comparison between EUS-FNA and surgical specimens. Curr Mol Med 2014;14:309-15. [Crossref] [PubMed]
- Ding B, Lou W, Xu L, et al. Analysis the prognostic values of solute carrier (SLC) family 39 genes in gastric cancer. Am J Transl Res 2019;11:486-98. [PubMed]
- Zhou H, Zhu Y, Qi H, et al. Evaluation of the prognostic values of solute carrier (SLC) family 39 genes for patients with lung adenocarcinoma. Aging (Albany NY) 2021;13:5312-31. [Crossref] [PubMed]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23-38. [Crossref] [PubMed]
- Maruyama R, Choudhury S, Kowalczyk A, et al. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet 2011;7:e1001369. [Crossref] [PubMed]
- Wang X, Li Y, Hu H, et al. Comprehensive analysis of gene expression and DNA methylation data identifies potential biomarkers and functional epigenetic modules for lung adenocarcinoma. Genet Mol Biol 2020;43:e20190164. [Crossref] [PubMed]
- Li P, Schille C, Schweizer E, et al. Mechanical Characteristics, In Vitro Degradation, Cytotoxicity, and Antibacterial Evaluation of Zn-4.0Ag Alloy as a Biodegradable Material. Int J Mol Sci 2018;19:755. [Crossref] [PubMed]
- Dimauro I, Antonioni A, Mercatelli N, et al. The early response of αB-crystallin to a single bout of aerobic exercise in mouse skeletal muscles depends upon fiber oxidative features. Redox Biol 2019;24:101183. [Crossref] [PubMed]
- Antonioni A, Dimauro I, Fantini C, et al. αB-crystallin response to a pro-oxidant non-cytotoxic environment in murine cardiac cells: An "in vitro" and "in vivo" study. Free Radic Biol Med 2020;152:301-12. [Crossref] [PubMed]
- Ohto-Fujita E, Hayasaki S, Atomi A, et al. Dynamic localization of αB-crystallin at the microtubule cytoskeleton network in beating heart cells. J Biochem 2020;168:125-37. [Crossref] [PubMed]
- Tang Q, Liu YF, Zhu XJ, et al. Expression and prognostic significance of the alpha B-crystallin gene in human hepatocellular carcinoma. Hum Pathol 2009;40:300-5. [Crossref] [PubMed]
- Chen P, Ji W, Liu FY, et al. Alpha-crystallins and tumorigenesis. Curr Mol Med 2012;12:1164-73. [Crossref] [PubMed]
- Liu XL, Guo KP, Ma F, et al. Expression profile of heat shock proteins in tissues and cells of lung adenocarcinoma. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2007;32:660-4. [PubMed]
- Choi S, Chen M, Cryns VL, et al. A nuclear phosphoinositide kinase complex regulates p53. Nat Cell Biol 2019;21:462-75. [Crossref] [PubMed]
- Klemenz R, Scheier B, Müller A, et al. Alpha B crystallin expression in response to hormone, oncogenes and stress. Verh Dtsch Ges Pathol 1994;78:34-5. [PubMed]
- Pinder SE, Balsitis M, Ellis IO, et al. The expression of alpha B-crystallin in epithelial tumours: a useful tumour marker? J Pathol 1994;174:209-15. [Crossref] [PubMed]
- Chen D, Cao G, Qiao C, et al. Alpha B-crystallin promotes the invasion and metastasis of gastric cancer via NF-κB-induced epithelial-mesenchymal transition. J Cell Mol Med 2018;22:3215-22. [Crossref] [PubMed]
- Zhu Z, Luo L, Xiang Q, et al. MiRNA-671-5p Promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell Death Dis 2020;11:949. [Crossref] [PubMed]
- Chatterjee S, Burns TF. Targeting Heat Shock Proteins in Cancer: A Promising Therapeutic Approach. Int J Mol Sci 2017;18:1978. [Crossref] [PubMed]
- Chatterjee S, Bhattacharya S, Socinski MA, et al. HSP90 inhibitors in lung cancer: promise still unfulfilled. Clin Adv Hematol Oncol 2016;14:346-56. [PubMed]
- Johnson JO, Gibbs JR, Megarbane A, et al. Exome sequencing reveals riboflavin transporter mutations as a cause of motor neuron disease. Brain 2012;135:2875-82. [Crossref] [PubMed]
- Yu Y, Wu A, Zhang Z, et al. Characterization of the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J Nutr Biochem 2013;24:1697-708. [Crossref] [PubMed]
- Wu L, Chaffee KG, Parker AS, et al. Zinc transporter genes and urological cancers: integrated analysis suggests a role for ZIP11 in bladder cancer. Tumour Biol 2015;36:7431-7. [Crossref] [PubMed]
- Chornokur G, Lin HY, Tyrer JP, et al. Common Genetic Variation In Cellular Transport Genes and Epithelial Ovarian Cancer (EOC) Risk. PLoS One 2015;10:e0128106. [Crossref] [PubMed]
- Barresi V, Valenti G, Spampinato G, et al. Transcriptome analysis reveals an altered expression profile of zinc transporters in colorectal cancer. J Cell Biochem 2018;119:9707-19. [Crossref] [PubMed]
- Liu L, Yang J, Wang C. Analysis of the prognostic significance of solute carrier (SLC) family 39 genes in breast cancer. Biosci Rep 2020;40:BSR20200764. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


