
Targeting chromosome 12q amplification in relapsed glioblastoma: the use of computational biological modeling to identify effective therapy—a case report
Introduction
Glioblastoma (GBM) is a lethal disease with a median survival of approximately one year for patients with MGMT-unmethylated disease and 18 months with MGMT-methylated cancers. Fewer than 5% survive 5 years. Following conventional therapy with surgery, chemoradiotherapy, adjuvant temozolomide and tumor treating fields (TTF), patients usually relapse with remarkably short survival. Typically, the relapsed GBM is marginally responsive to salvage therapies, such as lomustine, regorafenib, bevacizumab and other cytotoxic agents. In general, second-line treatments achieve significant radiographic responses in just 3–5% of cases. Hence, the discovery of effective therapy for this disease remains an urgent and unmet need.
Thus far, precision medicine based on next generation sequencing (NGS) alone has afforded little progress against GBM compared to other malignancies. Apparently, single agent oncogene-targeting yields little benefit against malignancies fueled by multiple signaling pathway aberrations. Additionally, tumor heterogeneity, transcriptional plasticity and glioma-mesenchymal transition constitute significant impediments. The blood brain barrier (BBB) also poses an obstacle to drug delivery. On the other hand, computational biological modeling (CBM), i.e., biosimulation, of multiple complex signaling pathways has recently emerged as a technique for integrating genomic information into effective treatment recommendations. A virtual avatar of a patient’s cancer generated from comprehensive genomic inputs permits interrogation of the disease network with regard to the impact size of various drug combinations on the hallmark behaviors of cancer. The feasibility of CBM to identify the likelihood of benefit from various GBM therapies has recently been demonstrated (1). Biosimulation of drug response demonstrates high positive and negative predictive value (~90%) for predicting clinical outcomes in GBM (2,3) and recently has received attention for GBM and other treatment-resistant diseases (4,5). In a population of 100 patients with GBM, biosimulation of treatment response was found to be strongly predictive of disease free survival (P=0.0266) and overall survival (P=0.0125), offering evidence that validates biosimulation (6). We report here a case of a patient with relapsed GBM for whom biosimulation led to a novel therapy which produced dramatic disease regression. We present the following article in accordance with the CARE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-2022-62/rc).
Case presentation
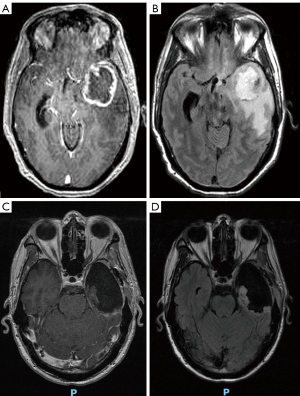
A 52-year-old man came to medical attention with 6 months of worsening mental status and personality changes. At presentation, MRI revealed a large L. cerebral tumor with vasogenic edema, local mass effect, and midline shift (Figure 1). Left frontotemporal craniotomy was performed for radical gross total resection utilizing a stereotactic computer-assisted, volumetric, intracranial procedure with frameless stereotaxis employing BrainLab™ (Brainlab AG, Munich, Germany) intraoperative navigation. Frozen section revealed high-grade glioma. Surgery was uneventful and the patient had a satisfactory post-operative MRI. He became more alert over the subsequent days in hospital and was discharged home without complications.
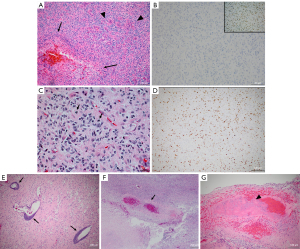
Pathology revealed GBM (WHO grade IV), extensive necrosis, thrombosis and hemorrhage, unusually brisk hypervascularity, abundant mitoses, and Ki-67 10–20% (Figure 2). The MGMT was unmethylated. Immunohistochemistry revealed GFAP positivity, IDH1/2 negativity, p53<10%, absence of 1p/19q co-deletion, or polysomy 7. Monosomy 10q was present.
Following surgery, the patient received conventional therapy with chemoradiotherapy (60 Gy + temozolomide 75 mg/m2 daily) and 6 cycles temozolomide (150 mg/m2 d 1–5 q 28 d). He declined TTF therapy. He entered a period of disease control that lasted for 8 months prior to the development of tumor relapse in the corpus callosum and the anterior horn of the contralateral R. ventricle.
The subject of this case report was not a study or clinical trial of any kind, and thus the application of Helsinki Declaration was not required. Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
NGS
PTEN and TERT promoter mutations, as well as 12q14-15 amplification with 20x copies of CDK4 and MDM2 was demonstrated (Tempus, Inc., Chicago, IL, USA).
Details of CBM and in silico biosimulation
The patient’s genomic abnormalities served as input for a novel computational biological model of cellular behavior constructed from a variety of data sources, including studies on cell receptors, signaling pathways, transcription factors and enzyme kinetics. The model was developed and validated using PubMed to generate protein network maps of patient-specific dysregulated pathways associated with hallmark behaviors of cancer (7-10). To ensure accuracy of computational simulation, published data were aggregated through manual scientific review. Simulation experiments and analyses using a dynamic representation of signaling and metabolic pathways were validated in cell line experiments to develop a predictive tumor model.
CBM includes representations of growth factor signaling, cell cycle regulation, tumor metabolism, oxidative stress, epigenetics, protein homeostasis, DNA damage repair, apoptosis, survival, angiogenesis, invasion and immune evasion. The current version of the model includes 3,765 genes and 29,181 functional interactions associated with cancer, including 286 kinases, 379 transcription factors, and 115 pathways which provide comprehensive coverage of the kinome, transcriptome, proteome and metabolome.
Creation of disease avatar using the CBM platform
Protein-protein interactions are simulated mathematically using Michaelis-Menten equations until the system reaches homeostasis and is considered to be in a control state. Subsequently, a virtual (i.e. in silico) disease model based on somatic gene mutations and gene copy number variations (CNVs) from the patient is used to generate phenotypic behaviors associated with malignancy. Specifically, the computational model derives a composite score of cell number from individual phenotype scores representing hallmark behaviors of cancer, including proliferation, survival, apoptotic blockade, genomic instability, invasion, angiogenesis and immune evasion. In this fashion, the impact of individual NGS results on patient-specific protein networks was computationally modeled.
Biosimulation of drug response
An agnostic drug library of 250 drugs was employed to model each drug singly and in 2- and 3-way combinations to interrogate the signaling network representing the patient’s disease avatar. The computational impact of drug simulation on the phenotypic behaviors of malignancy was assessed and ranked in a histogram representing anticipated drug efficacy.
Results of biosimulation
Biosimulation ranked the impact of various treatments against the composite disease phenotype (Figure 3). Efficacy was identified for nelfinavir, everolimus, palbociclib, leflunomide and selinexor and combinations of these agents. No efficacy was demonstrated for standard GBM salvage agents, including temozolomide, lomustine, bevacizumab and regorafenib. Theranostic associations from the model are shown in Table 1.
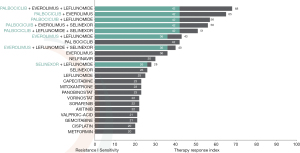
Table 1
| Gene | Pathway consequences | Recommended drug | References |
|---|---|---|---|
| PTEN-biallelic loss (mutation and monosomy 10) | Loss causes unrestrained PI3K signaling that generates high levels of PIP3 which activates AKT and triggers multiple drivers of malignant behavior | Nelfinavir inhibits HSP90 which effectively diminishes AKT activation | (11,12) |
| PTEN | Loss upregulates AKT signaling and enhances conversion of glutamine into pyrimidines through the de novo pyrimidine synthesis pathway. As a result, PTEN and DHODH are synthetic lethal partners | Leflunomide, a DHODH inhibitor, precipitates cell death | (13) |
| CDK4 amp | Amplification enhances phosphorylation of CCND1 which in turn phosphorylates RB1 to cause dissociation from its transcriptional target, E2F1, to promote entry into the cell cycle | CDK4/6 inhibitors result in tumor regression and a net reduction in tumor burden | (14-19) |
| MDM2 amp | Amplification results in p53 ubiquitination leading to increased proteasomal turnover resulting in genomic instability and apoptotic blockade | Nelfinavir blocks proteasomal degradation of p53 thus enhancing apoptosis by rescuing p53 | (20-22) |
CBM, computational biological modeling.
Treatment selection
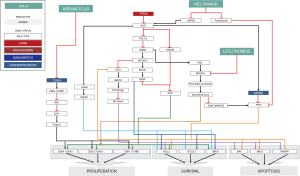
Combination therapy employing abemaciclib, nelfinavir and leflunomide was administered based on the biosimulation results. The mechanistic signaling pathway impact of genomic aberrations and treatment are shown in Figure 4. Abemaciclib was selected in preference to palbociclib because of superior BBB penetrance. The dosage of abemaciclib 150 mg once daily was reduced to half of the usual daily dosage because of downregulation of CYP3A4 by nelfinavir. Nelfinavir rather than everolimus was selected because of potential additive toxicities of everolimus and abemaciclib, and the absence of combinatorial phase IB experience with this combination. Nelfinavir dosage of 625 mg bid was chosen in preference to the usual single agent dosage of 1,250 mg bid because of the anticipated interaction with abemaciclib. Leflunomide was administered with a standard loading dose 100 mg daily ×3 day, followed by a maintenance dose 20 mg daily. The patient’s clinical status and blood counts and chemistries were monitored on a biweekly basis.
Response and tolerance of treatment
Serial MRI scans demonstrated immediate and substantial disease regression (Figure 5). Additionally, the patient noticed an improvement in cognitive functioning and regained the ability to conduct activities of daily living without supervision. Disease progression was identified by MRI at 6 months. Expected abemaciclib toxicities, including grade 2–3 leukopenia and grade 1–2 thrombocytopenia were observed. Diarrhea, hyperglycemia and lipodystrophy from nelfinavir did not occur. Protease inhibitors are known to suppress GLUT4 and may cause insulin resistance, hyperglycemia, dyslipidemia, lipodystrophy, hepatic steatosis and hepatomegaly by inhibiting the breakdown of SREBP in adipose and liver tissues (23). These toxicities could exacerbate the effects of corticosteroids in the neuro-oncology patient; however they were not observed at the relatively low dose of nelfinavir employed. Additionally, no dose limiting toxicities were observed from the use of leflunomide.
Discussion
The complexity and diversity of molecular aberrations in most cancers creates a daunting therapeutic challenge that has not been adequately addressed by targeting single oncogenes, nor by randomly combining drugs with different mechanisms of action and non-overlapping toxicities. Yet two decades ago, Hanahan and Weinberg espoused an integrated circuit model of the cell and foresaw the development of both comprehensive genomic profiling and computational biology to explain how specific genetic lesions reprogram the cell and induce the phenotypic “hallmarks of cancer” (24). The arrival of CBM to create an in silico disease avatar from comprehensive genomic inputs that can be interrogated with various drug combinations fulfills their prediction. As such, biosimulation provides a practical tool that addresses disease heterogeneity in the clinic and furthers the mission of making cancer treatment a “rational science”. In the case presented here, four driver pathways were simultaneously co-targeted with a novel drug triplet.
The 12q14-15 amplification constitutes a discreet chromosomal syndrome in this patient’s cancer as well as other malignancies across the cancer spectrum including soft tissue sarcomas (25,26), bladder cancer (27), neuroblastoma (28) and breast cancer (29). It is present in at least 10% of patients with GBM (30). After chromosome 7p amplification (18%), 12q13-15 is the second most frequently amplified genomic segment in GBM, followed by 4q12 (7%), and 1q32 (4%). These apparently non-random amplifications represent discreet subtypes of GBM defined by specific combinations of oncogenes that make up key driver mechanisms. When amplified, the sequence of genes on the amplicon defines oncogene neighborhoods that provide unique drivers of disease behavior.
Given the presence of CDK4 on the 12q14 amplicon, abemaciclib appears to be especially relevant for this group of glioma patients. Until a brain-penetrant MDM2 inhibitor can be identified, proteasomal inhibition with nelfinavir represents a precision medicine strategy to address the problem of MDM2 amplification causing p53 loss. Notably, the effect of nelfinavir on both AKT suppression and proteasomal degradation accomplishes downstream targeting of both disease drivers (PTEN and MDM2) simultaneously. Given the challenge of disabling concurrent CDK4 and MDM2 drivers arising from 12q amplification, the evaluation of abemaciclib and nelfinavir could be a useful strategy for controlling this commonly encountered subtype of GBM with an estimated incidence of 1,400 cases per year in the USA, as well as in other cancers with 12q amplification. Finally, PTEN loss is one of the most common abnormalities identified in GBM, suggesting that a synthetic lethal strategy with leflunomide may have broad potential against the PTEN-deficient GBM.
In addition to providing novel therapy described, biosimulation also suggested that selinexor is a potentially active drug. Given the availability of clinical trials evaluating selinexor in GBM, biosimulation can assist in selecting which clinical trials are most likely to demonstrate a therapeutic benefit for specific patients.
With attention to the administration of drugs that impact each other’s elimination, the benefits reported in this case were achieved with half doses of nelfinavir and abemaciclib. The observed leukopenia and thrombocytopenia characteristic of abemaciclib toxicity suggest that a therapeutic threshold concentration was achieved in spite of dose attenuation. The strategy also demonstrates that drug interactions can be exploited rather than shunned to derive substantial cost savings from combinations that rely on a single metabolic pathway for elimination.
The strength of the biosimulation approach is that it is able to model the consequences of dozens or hundreds of aberrant pathways to determine the impact of the patient’s genomic aberrations of on drug responsiveness. However, one limitation of the approach is that computational modeling requires comprehensive genomic information a large NGS panel with a sequencing depth sufficient to provide detailed copy number aberrations. As such, biosimulation awaits an evolution in clinical practice away from mutation-only analyses and limited gene panels which fall short of embracing the complexity and heterogeneity in cancer. Additionally, while initial evidence validating this artificial intelligence (AI) tool with regard to predicting disease free and overall survival has begun to emerge, a great deal more prospective validation will likely be necessary before this approach gains widespread acceptance.
An ocean of genomic information from the cancer research enterprise has brought an unprecedented insight into the mechanisms of malignancy and drug response. However, the task of accessing, digesting, and integrating specific knowledge relevant to all the elements of patients’ unique genomic profiles into optimal personalized treatment programs exceeds the practical limits of busy oncology practices. On the other hand, biosimulation of drug response brings AI to the clinic in a point-of-care tool that bridges the chasm between comprehensive molecular diagnosis and treatment selection. In the next chapter of the precision medicine story, the integration of molecular knowledge relevant to all unique aberrations in an individual’s cancer with therapeutic decision-making heralds a major development in oncology that transcends the one drug-one gene approach. Nearly three decades after the phrase was coined, what is meant by “translational medicine” is not simply a process of dedicating years of research to bringing a single treatment to bear against a particular oncogene, but can be a real-time, daily clinical routine of developing a molecular diagnoses for individual patients, modeling the consequences and impacts of various treatments, and then implementing the best options in the clinic. In this way, biosimulation has the potential to make precision medicine considerably more precise and in the process to bring improvements to disease control and survival.
Conclusions
CBM of multiple genomic abnormalities permits in silico modeling of a patient’s cancer and the signaling pathway aberrations responsible for disease behavior and drug resistance. As such CBM provides a deeper look at the mechanistic determinants of disease offering engineering-level insight into complex genomic information. At the same time, biosimulation of how various drug therapies interact with the individual patient’s disease network connects the insights of the molecular oncology with the relevant therapeutic vulnerabilities and necessities with an AI tool that provides stratified and actionable treatment information. As such biosimulation has the potential to extend the horizon of precision medicine beyond the current one gene-one drug paradigm to embrace the complexity and uniqueness of each patient’s cancer. For disease subtypes like 12q amplification, biosimulation also serves a discovery process of identifying novel drug combinations that may merit clinical trials testing and provide new impetus for the continued incremental advancement of personalized cancer therapy.
Acknowledgments
Funding: Cellworks Group Inc. (S. San Francisco, CA, USA) provided computational biology model testing for patients with internal funding.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-2022-62/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-2022-62/coif). MPC has accepted employment at Cellworks and provides consultation services to Exact Sciences and Caris Life Sciences, and reports speakers honoraria from Guardant health. A Pampana, AA, DAL, KGGR, ARRA, A Prakash, DS, LB, AK, SK have accepted employment at Cellworks. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The subject of this case report was not a study or clinical trial of any kind, and thus the application of Helsinki Declaration was not required. Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Castro M, Pampana A, Alam A, et al. Combination chemotherapy versus temozolomide for patients with methylated MGMT (m-MGMT) glioblastoma: results of computational biological modeling to predict the magnitude of treatment benefit. J Neurooncol 2021;153:393-402. [Crossref] [PubMed]
- Wen PY, Watson D, Kapoor S, et al. Superior therapy response predictions for patients with glioblastoma (GBM) using Cellworks Singula: MyCare-009-03. J Clin Oncol 2020;38:2519. [Crossref]
- Ahluwalia MS, Watson D, Kapoor S, et al. Superior therapy response predictions for patients with low-grade glioma (LGG) using Cellworks Singula: MyCare-009-04. J Clin Oncol 2020;38:2569. [Crossref]
- Deisboeck TS, Zhang L, Yoon J, et al. In silico cancer modeling: is it ready for prime time? Nat Clin Pract Oncol 2009;6:34-42. [Crossref] [PubMed]
- Rahman R, Trippa L, Alden S, et al. Prediction of Outcomes with a Computational Biology Model in Newly Diagnosed Glioblastoma Patients Treated with Radiation Therapy and Temozolomide. Int J Radiat Oncol Biol Phys 2020;108:716-24. [Crossref] [PubMed]
- Wen PY, Castro M, Watson D, et al. Superior overall survival (OS) and disease-free survival (DFS) predictions for patients with glioblastoma multiforme (GBM) using Cellworks Singula: myCare-022-03. J Clin Oncol 2021;39:2017. [Crossref]
- Drusbosky L, Medina C, Martuscello R, et al. Computational drug treatment simulations on projections of dysregulated protein networks derived from the myelodysplastic mutanome match clinical response in patients. Leuk Res 2017;52:1-7. [Crossref] [PubMed]
- Drusbosky LM, Cogle CR. Computational Modeling and Treatment Identification in the Myelodysplastic Syndromes. Curr Hematol Malig Rep 2017;12:478-83. [Crossref] [PubMed]
- Doudican NA, Kumar A, Singh NK, et al. Personalization of cancer treatment using predictive simulation. J Transl Med 2015;13:43. [Crossref] [PubMed]
- Pingle SC, Sultana Z, Pastorino S, et al. In silico modeling predicts drug sensitivity of patient-derived cancer cells. J Transl Med 2014;12:128. [Crossref] [PubMed]
- Georgescu MM. PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control. Genes Cancer 2010;1:1170-7. [Crossref] [PubMed]
- Gupta AK, Li B, Cerniglia GJ, et al. The HIV protease inhibitor nelfinavir downregulates Akt phosphorylation by inhibiting proteasomal activity and inducing the unfolded protein response. Neoplasia 2007;9:271-8. [Crossref] [PubMed]
- Mathur D, Stratikopoulos E, Ozturk S, et al. PTEN Regulates Glutamine Flux to Pyrimidine Synthesis and Sensitivity to Dihydroorotate Dehydrogenase Inhibition. Cancer Discov 2017;7:380-90. [Crossref] [PubMed]
- Ezhevsky SA, Nagahara H, Vocero-Akbani AM, et al. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci U S A 1997;94:10699-704. [Crossref] [PubMed]
- Rubin SM, Gall AL, Zheng N, et al. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 2005;123:1093-106. [Crossref] [PubMed]
- Dong Y, Sui L, Sugimoto K, et al. Cyclin D1-CDK4 complex, a possible critical factor for cell proliferation and prognosis in laryngeal squamous cell carcinomas. Int J Cancer 2001;95:209-15. [Crossref] [PubMed]
- Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3:1427-38. [Crossref] [PubMed]
- Logan JE, Mostofizadeh N, Desai AJ, et al. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity. Anticancer Res 2013;33:2997-3004. [PubMed]
- Huang S, Ye H, Guo W, et al. CDK4/6 inhibitor suppresses gastric cancer with CDKN2A mutation. Int J Clin Exp Med 2015;8:11692-700. [PubMed]
- Bono C, Karlin L, Harel S, et al. The human immunodeficiency virus-1 protease inhibitor nelfinavir impairs proteasome activity and inhibits the proliferation of multiple myeloma cells in vitro and in vivo. Haematologica 2012;97:1101-9. [Crossref] [PubMed]
- Hamel FG, Fawcett J, Tsui BT, et al. Effect of nelfinavir on insulin metabolism, proteasome activity and protein degradation in HepG2 cells. Diabetes Obes Metab 2006;8:661-8. [Crossref] [PubMed]
- Kraus M, Bader J, Overkleeft H, et al. Nelfinavir augments proteasome inhibition by bortezomib in myeloma cells and overcomes bortezomib and carfilzomib resistance. Blood Cancer J 2013;3:e103. [Crossref] [PubMed]
- Hui DY. Effects of HIV protease inhibitor therapy on lipid metabolism. Prog Lipid Res 2003;42:81-92. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Kyriazoglou AI, Vieira J, Dimitriadis E, et al. 12q amplification defines a subtype of extraskeletal osteosarcoma with good prognosis that is the soft tissue homologue of parosteal osteosarcoma. Cancer Genet 2012;205:332-6. [Crossref] [PubMed]
- Creytens D, Van Gorp J, Speel EJ, et al. Characterization of the 12q amplicons in lipomatous soft tissue tumors by multiplex ligation-dependent probe amplification-based copy number analysis. Anticancer Res 2015;35:1835-42. [PubMed]
- Simon R, Struckmann K, Schraml P, et al. Amplification pattern of 12q13-q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene 2002;21:2476-83. [Crossref] [PubMed]
- Guimier A, Ferrand S, Pierron G, et al. Clinical characteristics and outcome of patients with neuroblastoma presenting genomic amplification of loci other than MYCN. PLoS One 2014;9:e101990. [Crossref] [PubMed]
- Gómez-Miragaya J, Díaz-Navarro A, Tonda R, et al. Chromosome 12p Amplification in Triple-Negative/BRCA1-Mutated Breast Cancer Associates with Emergence of Docetaxel Resistance and Carboplatin Sensitivity. Cancer Res 2019;79:4258-70. [Crossref] [PubMed]
- González-Tablas M, Crespo I, Vital AL, et al. Prognostic stratification of adult primary glioblastoma multiforme patients based on their tumor gene amplification profiles. Oncotarget 2018;9:28083-102. [Crossref] [PubMed]



