
High expression of aurora kinase B predicts poor prognosis in hepatocellular carcinoma after curative surgery and its effects on the tumor microenvironment
Introduction
Primary liver cancer has high morbidity and mortality, causing an estimated 830,000 deaths globally in 2020 and representing 8.3% of all cancer fatalities (1). The most frequent subtype of primary liver cancer is hepatocellular carcinoma (HCC), which is attributable to around 75–85% of all primary liver cancer cases (2). Despite the availability of numerous therapies such as chemotherapy, immunotherapy, and molecularly targeted therapy, surgical intervention remains the primary therapeutic option for patients with HCC (3,4). However, the recurrence rate of HCC is high, and the long-term prognosis remains poor. Currently, HCC still lacks effective prognostic markers. Therefore, finding novel biomarkers and therapeutic targets, as well as investigating the pathophysiology of HCC, is of the utmost significance since it has the potential to enhance both the prognosis and the treatment of the disease.
Aurora kinases (AURKs) belong to the family of serine/threonine kinases, which has three members (AURKA, AURKB, and AURKC) (5). AURKs are important regulators of the cell cycle, of which AURKA and AURKB are the most abundant isoform in cancer (6). Overexpression of AURKs in tumors has been shown to trigger genomic instability that leads to tumor development, invasion, and metastasis (7). In particular, numerous studies have shown the AURKB expression level is elevated in malignancies, and this is linked to the onset and advancement of cancer (8-10). As a result of its involvement in cancer cell proliferation, epithelial-mesenchymal transition (EMT), apoptosis, metastasis, and the self-renewal of cancer stem cells, AURKB contributes to the progression of tumors (11-15). In addition, it is known to be implicated in the modulation of several signaling pathways, particularly the p70S6K/RPL15, MAPK/ERK, NF-κB pathways, and other signaling pathways (16-18). Furthermore, the expression of AURKB is identified as a prognostic biomarker in neuroblastoma, clear cell renal carcinoma (ccRCC), and pediatric acute lymphoblastic leukemia (10,19,20). The results of these investigations suggested AURKB could perform an important part in oncogenesis. While several studies have demonstrated that AURKB is a promising prognostic biomarker in HCC (21-25), neither its function in HCC nor its possible molecular basis is thoroughly studied at this time, especially for microvascular invasion (MVI) and the tumor immune microenvironment (TIME).
In this research, we evaluated the expression level, diagnostic role, prognostic value, and the possible association of the expression of AURKB with clinical and pathological features, and its relationship with immune cell infiltration and immune checkpoints was investigated. In addition, we identified AURKB-related differentially expressed genes (DEGs) and explored the putative molecular mechanisms in the advancement of HCC by functional enrichment analysis. Our findings illustrated AURKB was substantially up-modulated in HCC and could be a viable diagnostic biological marker for the disease. Additionally, AURKB had an important impact on TIME. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4798/rc).
Methods
Acquisition of public data
The gene expression, clinicopathological information, and matching survival data of HCC patients were obtained from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) and Gene Expression Omnibus (GEO). The RNA-sequencing data of 351 HCC patients were included in the TCGA-LIHC cohort, and the transcripts per million (TPM) approach was utilized for gene expression quantification. In addition, microarray expression data from 221 patients with HCC were also downloaded from the GSE14520 cohort. HCC patients who had incomplete clinical/survival data were excluded from this study.
Patients and clinical samples
In total, this study enrolled 262 patients diagnosed with HCC and treated surgically at the Third Affiliated Hospital of Naval Military Medical University between 2013 and 2015. The term “over survival” (OS) was used to refer to the duration of time that passed between the patients’ surgery and the date of their death or their last follow-up. Calculation of the recurrence-free survival (RFS) began on the date of the surgical procedure until the initial diagnosis of recurrence or death, and patients were monitored until June 2021. The principles highlighted in the Declaration of Helsinki (as revised in 2013) were adhered to throughout this research. The study was approved by the Ethics Committee of Third Affiliated Hospital of Naval Military Medical University and informed consent was taken from all the patients.
Immunohistochemistry (IHC) staining analysis
For IHC analysis, HCC tissues were surgically removed, and 4% formalin was utilized to fix them before embedding in paraffin blocks. Following antigen retrieval via microwave heating, slides were treated throughout the night with the primary anti-AURKB antibody (diluted at 1:200, ab2254, Abcam, UK). Hematoxylin was then utilized as a counterstain after the specimens had been stained with 3',3-diaminobenzidine tetrahydrochloride (DAB), and following alcohol dehydration, xylene vitrification, and neutral gum sealing, a total of four random views of each sample were examined microscopically at a magnification of ×400. For AURKB expression evaluation, the proportion of positive cells in each view was used to derive the score, with a positive cell number ≤25% =1, 26–50% =2, 51–75% =3, and 76–100% =4. In addition, for the staining intensity score; negative =1, weak =2, moderate =3, and strong =4. The composite scores of 1–4 (+), 5–8 (++), 9–12 (+++), and 13–16 (++++), were obtained using the formula; composite score = percentage score of positive cells × intensity grade. According to the intensity composite score, low levels of AURKB expression were indicated by scores between 1 and 8 (+ and ++), whereas high levels were indicated by values between 9 and 16 (+++ and ++++). Examination of the staining was carried out by two pathologists independently.
Survival analysis and nomogram construction
All patients whose follow-up time was ≥1 month were considered eligible for inclusion in this research. Patients were classified into two groups (low expression and high expression) depending on the optimum thresholds of the gene expression level using the “surv cutpoint” function included in the “survminer” R package. The log-rank test was employed to examine differences (variations) in survival rates. To discover independent prognostic variables in HCC, both univariate and multivariate Cox analyses were conducted, and by using the “rms” R package, a nomogram that considered prognostic risk variables and AURKB expression was developed. Both discrimination and calibration were used to verify the ability of the nomogram to accurately predict patient survival.
Correlation of AURKB expression with TIME
The CIBERSORT method and the LM22 signature matrix were utilized to identify immune cell infiltration in HCC patients by estimating the percentages of various types of immune cells (26). LM22 is a signature matrix that can differentiate different immune cells in an accurate manner, such as different types of naïve and memory B cells, natural killer (NK) cells, plasma cells, T cells, and myeloid subsets. In addition, we investigated the relationships between AURKB expression and T cell markers (CD3D, CD8A, CD4, and FOXP3), macrophage markers (CD68, CD86, CD163, and GZMB), and immune checkpoints (PDCD1, CD274, CTLA4, and LAG3).
Screening of DEGs and analysis of functional enrichment
The DEGs related to AURKB in HCC tissues were analyzed using the R package “edgeR” (27). False discovery rate (FDR) adjusted P<0.05 and Log2 fold change >1.0 were established as the thresholds, and the findings were displayed by a volcano plot. The “clusterProfiler” and “enrichplot” packages in R software were employed to analyze Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, which identified possible mechanisms linked to DEGs (28,29). Significant enrichment was considered to exist when the P value was <0.05.
Statistical analysis
The χ2 test or the Fisher exact test, and the t-test were used, respectively, to compare categorical and continuous variables following a normal distribution, and continuous variables exhibiting skewed distributions were evaluated utilizing the Mann-Whitney U test. Additionally, the log-rank test was conducted to determine whether there were remarkable variations in survival rates across the two groups. Using linear regression analysis, we determined the link that exists between the expression of AURKB and that of other genes. Analyses of statistical data were conducted using R software, and a statistically significant variation was judged to exist when the two-tailed P value was <0.05.
Results
Clinical features of HCC patients
Firstly, 262 clinical samples were acquired from HCC patients who received surgical treatment at our hospital from 2013 to 2015. As depicted in Table 1, detailed parameters included age, gender, total bilirubin (TBIL), aspartate aminotransferase (AST), alanine aminotransferase (ALT), hepatitis B virus (HBV)-DNA, Child-Pugh score, Edmondson-Steiner grade, tumor node metastasis (TNM) stage, Barcelona clinic liver cancer (BCLC) stage, tumor size, liver cirrhosis, alpha-fetoprotein (AFP), tumor number, MVI, and satellite lesions. In addition, the clinical data of HCC patients from TCGA-LIHC and the GSE14520 cohort were also described.
Table 1
| Variables | Hospital samples (n=262) |
GSE14520 (n=221) |
TCGA-LIHC (n=351) |
|---|---|---|---|
| Age | |||
| <60 years | 103 | 178 | 157 |
| ≥60 years | 159 | 43 | 194 |
| Gender | |||
| Female | 28 | 30 | 110 |
| Male | 234 | 191 | 241 |
| TBIL | |||
| ≤17.1 μmol/L | 226 | N/A | N/A |
| >17.1 μmol/L | 36 | N/A | N/A |
| AST | |||
| ≤40 U/L | 113 | N/A | N/A |
| >40 U/L | 149 | N/A | N/A |
| ALT | |||
| ≤44 U/L | 151 | N/A | N/A |
| >44 U/L | 111 | N/A | N/A |
| HBV-DNA | |||
| <2,000 IU/mL | 112 | N/A | N/A |
| ≥2,000 IU/mL | 150 | N/A | N/A |
| Child-Pugh score | |||
| A | 238 | N/A | N/A |
| B/C | 24 | N/A | N/A |
| Edmondson-Steiner grade | |||
| I–II | 56 | N/A | N/A |
| III–IV | 206 | N/A | N/A |
| TNM stage | |||
| I/II | N/A | 170 | 246 |
| III/IV | N/A | 49 | 83 |
| BCLC stage | |||
| 0/A | N/A | 168 | N/A |
| B/C | N/A | 51 | N/A |
| Tumor size | |||
| ≤5 cm | 121 | 140 | N/A |
| >5 cm | 141 | 80 | N/A |
| Liver cirrhosis | |||
| No | 72 | N/A | N/A |
| Yes | 190 | N/A | N/A |
| AFP | |||
| ≤400 ng/mL | 83 | 118 | N/A |
| >400 ng/mL | 179 | 100 | N/A |
| Tumor number | |||
| 1 | 195 | N/A | N/A |
| >1 | 67 | N/A | N/A |
| MVI | |||
| No | 139 | N/A | N/A |
| Yes | 123 | N/A | N/A |
| Satellite lesions | |||
| No | 62 | N/A | N/A |
| Yes | 200 | N/A | N/A |
HCC, hepatocellular carcinoma; TCGA-LIHC, The Cancer Genome Atlas-Liver Hepatocellular Carcinoma; TBIL, total bilirubin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; HBV-DNA, hepatitis B virus-DNA; TNM, tumor node metastasis; BCLC, Barcelona clinic liver cancer; AFP, alpha fetoprotein; MVI, microvascular invasion; N/A, not applicable.
Expression and diagnostic function of AURKB in HCC
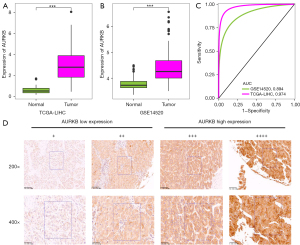
As per the results of the TCGA-LIHC cohort, the mRNA expression level of AURKB was substantially upmodulated in HCC tissues in contrast with the adjoining normal liver (P<0.001, Figure 1A). This result was confirmed in the GSE14520 HCC cohort (Figure 1B). In addition, the area under the curve (AUC) of AURKB in the TCGA-LIHC and GSE14520 cohorts were 0.974 and 0.894 correspondingly, which showed a high expression of AURKB had good diagnostic potential in HCC (Figure 1C). Additionally, the protein expression of AURKB was evaluated by IHC staining of tissues from our hospital HCC cohort, which allowed for the 262 HCC patients to be categorized into two categories; an AURKB low-expression (low-AURKB) (n=149) group and an AURKB high-expression (high-AURKB) (n=113) group (Figure 1D). These findings illustrate AURKB is considerably upregulated in HCC and has good diagnostic potential.
Analysis of the relationship between clinical features and the expression of AURKB in HCC
We next determined whether there was a link between the expression of AURKB and the clinical features of HCC patients. In our clinical samples (Table 2), both the low- and high-expression groups did not vary significantly in terms of gender, age, TBIL, ALT, and AST levels, as well as HBV-DNA. Low-AURKB group patients exhibited a greater likelihood of having a lower Child-Pugh score (P=0.026) and Edmondson-Steiner grade (P<0.001), while those belonging to the AURKB-high group had a larger probability to have a higher incidence of MVI as opposed to patients in the AURKB-low group (P<0.001). However, tumor size, tumor number, liver cirrhosis, AFP, and satellite lesions showed no significant variation across the two groups.
Table 2
| Variables | Low-AURKB, n (%) |
High-AURKB, n (%) |
P |
|---|---|---|---|
| Age | 0.121 | ||
| <60 years | 52 (34.9) | 51 (45.1) | |
| ≥60 years | 97 (65.1) | 62 (54.9) | |
| Gender | 0.149 | ||
| Female | 20 (13.4) | 8 (7.08) | |
| Male | 129 (86.6) | 105 (92.9) | |
| TBIL | 0.992 | ||
| ≤17.1 μmol/L | 128 (85.9) | 98 (86.7) | |
| >17.1 μmol/L | 21 (14.1) | 15 (13.3) | |
| AST | 0.187 | ||
| ≤40 U/L | 70 (47.0) | 43 (38.1) | |
| >40 U/L | 79 (53.0) | 70 (61.9) | |
| ALT | 0.360 | ||
| ≤44 U/L | 90 (60.4) | 61 (54.0) | |
| >44 U/L | 59 (39.6) | 52 (46.0) | |
| HBV-DNA | 0.649 | ||
| <2,000 IU/mL | 66 (44.3) | 46 (40.7) | |
| ≥2,000 IU/mL | 83 (55.7) | 67 (59.3) | |
| Child-Pugh score | 0.026 | ||
| A | 141 (94.6) | 97 (85.8) | |
| B/C | 8 (5.37) | 16 (14.2) | |
| Edmondson-Steiner grade | <0.001 | ||
| I–II | 48 (32.2) | 8 (7.08) | |
| III–IV | 101 (67.8) | 105 (92.9) | |
| Tumor size | 1.000 | ||
| ≤5 cm | 69 (46.3) | 52 (46.0) | |
| >5 cm | 80 (53.7) | 61 (54.0) | |
| Tumor number | 1.000 | ||
| 1 | 111 (74.5) | 84 (74.3) | |
| >1 | 38 (25.5) | 29 (25.7) | |
| Liver cirrhosis | 0.067 | ||
| No | 48 (32.2) | 24 (21.2) | |
| Yes | 101 (67.8) | 89 (78.8) | |
| AFP | 0.648 | ||
| ≤400 ng/mL | 45 (30.2) | 38 (33.6) | |
| >400 ng/mL | 104 (69.8) | 75 (66.4) | |
| MVI | <0.001 | ||
| No | 101 (67.8) | 38 (33.6) | |
| Yes | 48 (32.2) | 75 (66.4) | |
| Satellite lesions | 0.511 | ||
| No | 38 (25.5) | 24 (21.2) | |
| Yes | 111 (74.5) | 89 (78.8) | |
| Recurrence | <0.001 | ||
| No | 89 (59.7) | 26 (23.0) | |
| Yes | 60 (40.3) | 87 (77.0) | |
AURKB, aurora kinase B; HCC, hepatocellular carcinoma; TBIL, total bilirubin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; HBV-DNA, hepatitis B virus-DNA; AFP, alpha-fetoprotein; MVI, microvascular invasion.
Prognostic value of AURKB in HCC
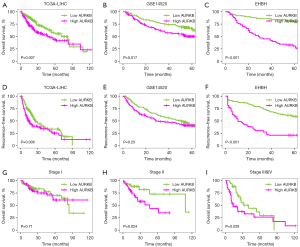
Further investigation into the prognostic significance of AURKB in HCC was conducted by Kaplan-Meier analysis. As per the findings, individuals with HCC who had an elevated expression level of AURKB had an unfavorable OS rate in the TCGA-LIHC cohort (P=0.007, Figure 2A), and in the GSE14520 cohort (P=0.017, Figure 2B). These findings were then verified using the information obtained from our clinical samples (P<0.001, Figure 2C). In addition, we found HCC patients with low-expression AURKB exhibited substantially better RFS than high-expression patients in the TCGA-LIHC and our clinical cohort, while no significant variations were seen in the GSE14520 cohort (Figure 2D-2F). As HCC is heterogeneous, we analyzed the prognostic significance of AURKB among HCC patients with different tumor TNM stages, and the findings illustrated elevated expression levels of AURKB predicted an unfavorable OS in stages II and III but not in stage I (Figure 2G-2I). These results showed that AURKB overexpression was considerably linked to dismal OS.
Establishment of a nomogram for OS premised on AURKB expression
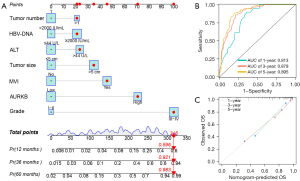
Following adjustment for other prognostic markers, univariate and multivariate Cox analyses revealed ALT, HBV-DNA, tumor size, tumor number, MVI, Edmondson-Steiner grade, and AURKB expression independently served as indicators of poor OS in patients with HCC (Table 3). We then developed a nomogram based on these risk factors. Figure 3A demonstrates the Edmondson-Steiner grade contributed more risk points (0–100) in contrast with AURKB expression, which was in line with the outcomes of the multivariate Cox regression analysis. The nomogram for predicting OS had a C-index of 0.794, and at 1, 3, and 5 years, the AUC of the receiver operating characteristic (ROC) curve for the HCC prognostic model was 0.813, 0.879, and 0.895, correspondingly (Figure 3B). As per the calibration plots, performance of the prediction using the nomogram was satisfactory in contrast with the likelihood of HCC patient survival (Figure 3C).
Table 3
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | 1.26 (0.64–2.5) | 0.500 | 1.00 (0.49–2.05) | 0.993 | |
| Age | 0.93 (0.64–1.36) | 0.723 | 1.34 (0.89–2.02) | 0.166 | |
| TBIL | 0.89 (0.51–1.56) | 0.685 | 0.95 (0.53–1.7) | 0.874 | |
| ALT | 1.21 (0.83–1.75) | 0.317 | 1.65 (1.09–2.52) | 0.019 | |
| AST | 1.44 (0.99–2.11) | 0.060 | 0.95 (0.63–1.44) | 0.814 | |
| HBV-DNA | 1.41 (0.97–2.07) | 0.075 | 1.58 (1.03–2.43) | 0.036 | |
| Child-Pugh score | 2.02 (1.17–3.47) | 0.012 | 1.54 (0.83–2.85) | 0.170 | |
| AFP | 1.55 (1.01–2.36) | 0.043 | 1.13 (0.7–1.83) | 0.622 | |
| Tumor size | 1.88 (1.28–2.76) | 0.001 | 1.99 (1.3–3.04) | 0.002 | |
| Tumor number | 1.31 (0.87–1.96) | 0.198 | 1.6 (1.04–2.48) | 0.033 | |
| Satellite lesions | 0.96 (0.63–1.48) | 0.864 | 0.82 (0.51–1.33) | 0.424 | |
| MVI | 4.41 (2.93–6.65) | <0.001 | 1.93 (1.2–3.11) | 0.007 | |
| Edmondson-Steiner grade | 10.83 (3.98–29.44) | <0.001 | 5.58 (1.98–15.75) | 0.001 | |
| Liver cirrhosis | 1.06 (0.7–1.61) | 0.783 | 0.77 (0.49–1.23) | 0.276 | |
| AURKB | 5.38 (3.54–8.17) | <0.001 | 3.63 (2.24–5.86) | <0.001 | |
AURKB, aurora kinase B; HCC, hepatocellular carcinoma; TBIL, total bilirubin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; HBV-DNA, hepatitis B virus-DNA; AFP, alpha-fetoprotein; MVI, microvascular invasion; HR, hazard ratio; CI, confidence interval.
Association between AURKB expression and immune microenvironment
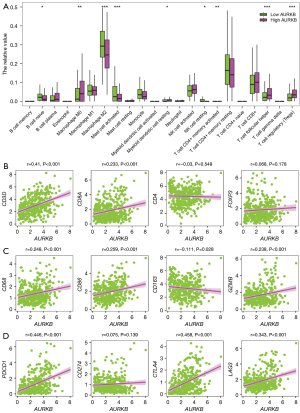
CIBERSORT was utilized for evaluation of the percentages of 22 immune cells within the TCGA-LIHC cohort, which allowed for assessment of the impact of AURKB on the immune microenvironment. As displayed in Figure 4A, the infiltrating levels of naive B cells, M2 macrophages, activated mast cells, and resting NK cells were decreased in the high-AURKB group, while M0 macrophages, regulatory T cells (Treg), T follicular helper cells (Tfh), and resting myeloid dendritic cells were increased. In addition, AURKB had a significantly positive link to the expression of CD3D (T cells marker) and CD8A (CD8+ T cells marker), and no significant association with CD4 (CD4+ T cells marker) and FOXP3 (Treg marker) (Figure 4B). The expression was also significantly positive with the expression of CD68 (macrophage marker), CD86 (M1 macrophage marker), and GZMB (cytotoxic marker), and negative with CD163 (M2 macrophage marker) (Figure 4C). Furthermore, analysis of the link between the AURKB and immune checkpoints was carried out to determine whether it could accurately predict the outcome of immunotherapy treatments, and our data showed its expression was positively linked to the expression of PDCD1, CD274, LAG3, and CTLA4 (Figure 4D).
Determination of DEGs linked to AURKB and functional analysis in HCC
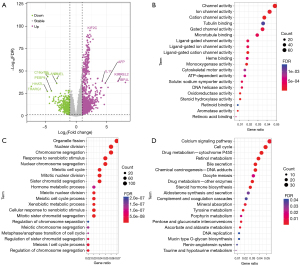
As illustrated in Figure 5A, we identified DEGs between the AURKB low- and high-expression categories in HCC, and the results show 1,194 genes were substantially up-modulated in the AURKB high-expression category and 502 genes were remarkably down-modulated. Additionally, we explored the potential biological processes and signaling pathways of AURKB-related DEGs implicated in HCC. The molecular functions of these DEGs revealed they were predominantly involved in channel activity, monooxygenase activity, heme binding, DNA helicase activity, oxidoreductase activity, and steroid hydroxylase activity (Figure 5B). The results of biological processes analysis illustrated predominant enrichment of DEGs in chromosome segregation, nuclear division, meiotic cell cycle, xenobiotic metabolic process, and regulation of chromosome separation (Figure 5C). Finally, KEGG pathway enrichment analysis illustrated considerable enrichment of 20 signaling pathways including the calcium signaling pathway, cell cycle, bile secretion, retinol metabolism, tyrosine metabolism, drug metabolism-cytochrome P450, and DNA replication (Figure 5D).
Discussion
Research suggests AURKB is an oncogene in gastric cancer, ccRCC, and lung adenocarcinoma, and is strongly elevated in several other malignancies (10,30,31). However, the predictive significance of AURKB and its possible mechanism, have not yet been completely investigated in HCC, particularly in terms of MVI and the TIME. Our results showed the level of mRNA expression of AURKB in HCC tissues was remarkably elevated as opposed to normal liver tissue samples, and the early identification and diagnosis of HCC patients are critical elements in their overall therapy and prognosis. We also discovered AURKB had a strong diagnostic performance for HCC, which suggested it may be a new biological marker for the clinical diagnosis of the disease.
Since the AURKB expression level was remarkably elevated in HCC, we evaluated its clinical significance by studying its association with clinical and pathological parameters. Our findings highlighted AURKB expression was linked to Child-Pugh score, MVI, Edmondson-Steiner grade, and tumor recurrence in clinical samples but did not associate with tumor size and tumor number. These results suggested AURKB may affect the survival prognosis of HCC through MVI and tumor recurrence. Recently, several research reports confirmed MVI as a high-risk indicator for post-surgical recurrence and metastasis of HCC, and that is has an important impact on long-term survival prognosis (32,33). As invasion and metastasis are two of the most important biological features of HCC, and because the microvascular system is the primary pathway via which HCC undergoes metastasis, MVI may be used to predict the tendency of tumor metastasis. Patients diagnosed with HCC who also had MVI exhibited a much more dismal prognosis in contrast with those who did not (34). We discovered 46.9% (123/262) of patients in our clinical samples had MVI, and these exhibited considerably elevated expression levels of AURKB. In addition, the expression of AURKB had a substantial correlation, not only with the pathological grade, but also with the recurrence of tumors. These data illustrate AURKB has important clinical significance in HCC.
We next determined the influence of AURKB expression on the survival prognosis of HCC patients, and as per the findings of our Kaplan-Meier analysis, those with elevated AURKB expression levels exhibited considerably more unfavorable RFS and OS status. Nonetheless, additional subgroup analysis illustrated that high-AURKB expression was an indicator of a negative prognosis only for patients who were in the middle or advanced stages of the disease and not those in the early stage. We took into consideration the possibility that the analysis was influenced by several additional prognostic indicators or the small sample size. Subsequently, the findings of univariate and multivariate Cox analysis illustrated ALT, HBV-DNA, tumor size, tumor number, MVI, Edmondson-Steiner grade, and AURKB expression were independent clinical factors of OS in HCC. Finally, a nomogram was generated using data on AURKB expression levels and other clinical risk factors to predict the prognosis of HCC patients, and as per the findings of our study, AURKB had significant predictive value, demonstrating its potential for use as a novel biological marker.
Research has shown the TIME performs an essential role in tumor onset and progression (35,36). It has been hypothesized that immune cells infiltrating tumors have a critical function in the tumor microenvironment (TME), and our findings illustrated low-AURKB patients had elevated levels of naive B cells, M2 macrophages, resting NK cells, and activated mast cells when contrasted with high-AURKB patients. Moreover, immunosuppressive Treg cells were significantly up-regulated in high-AURKB patients. Treg suppress the activities of CD4+ and CD8+ effector T cells as well as NK cells via a variety of mechanisms such as the release of immunosuppressive cytokines and the synthesis of cytolytic proteins. Subsequently, we also found AURKB was significantly positive with the expression of CD8A (CD8+ T cells marker), CD86 (M1 macrophage marker), and GZMB (cytotoxic marker), and negative with CD163 (M2 macrophage marker). Finally, we analyzed the potential of AURKB to make predictions about immunotherapeutic responsiveness and performed a correlation analysis between it and immune checkpoints. Based on our findings, a positive link exists between the levels of expression of AURKB and PDCD1, CD274, CTLA4, and LAG3. These findings illustrated patients with high-AURKB expression levels had a greater proportion of immunosuppressive cells, which suppress anti-tumor immune responses and results in a dismal prognosis.
In this study, we evaluated DEGs associated with AURKB and discovered that 1,194 genes were substantially positively related, and 502 genes were inversely related. We then conducted a functional enrichment analysis to elucidate the molecular mechanisms of AURKB-related DEGs in HCC. The outcomes of biological processes analysis showed a predominant enrichment of these DEGs in the meiotic cell cycle, chromosome segregation, xenobiotic metabolic process, nuclear division, and regulation of chromosome separation. As per the findings of the KEGG pathway enrichment analysis, there were 20 substantially enriched signaling pathways including the calcium signaling pathway, bile secretion, cell cycle, retinol metabolism, tyrosine metabolism, drug metabolism-cytochrome P450, and DNA replication. It is important to better define the basic processes through which AURKB enhances liver carcinogenesis in the future, given there has been no study addressing the carcinogenesis of AURKB through the previously mentioned pathways in HCC until now.
In summary, a comparison of HCC tissues with normal liver tissues revealed the expression level of AURKB was considerably elevated in the former. Overexpression of AURKB was associated with Edmondson-Steiner grade, MVI, and HCC recurrence, and independently served as a predictor of poor prognosis for HCC patients. Furthermore, AURKB expression was significantly associated with the immune microenvironment, especially for CD8+ T cells, macrophages, and immune checkpoints. Finally, AURKB was notably related to DNA replication, cell cycle, chromosome segregation, and nuclear division in HCC. However, more research is required to elucidate the function of AURKB at the molecular level and its significance in HCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4798/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4798/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4798/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Third Affiliated Hospital of Naval Military Medical University and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Liu CY, Chen KF, Chen PJ. Treatment of Liver Cancer. Cold Spring Harb Perspect Med 2015;5:a021535. [Crossref] [PubMed]
- Borah NA, Reddy MM. Aurora Kinase B Inhibition: A Potential Therapeutic Strategy for Cancer. Molecules 2021;26:1981. [Crossref] [PubMed]
- Du R, Huang C, Liu K, et al. Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy. Mol Cancer 2021;20:15. [Crossref] [PubMed]
- Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer 2005;5:42-50. [Crossref] [PubMed]
- Bertran-Alamillo J, Cattan V, Schoumacher M, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun 2019;10:1812. [Crossref] [PubMed]
- Zhu Q, Ding L, Zi Z, et al. Viral-Mediated AURKB Cleavage Promotes Cell Segregation and Tumorigenesis. Cell Rep 2019;26:3657-3671.e5. [Crossref] [PubMed]
- Wan B, Huang Y, Liu B, et al. AURKB: a promising biomarker in clear cell renal cell carcinoma. PeerJ 2019;7:e7718. [Crossref] [PubMed]
- Wang Z, Yu Z, Wang GH, et al. AURKB Promotes the Metastasis of Gastric Cancer, Possibly by Inducing EMT. Cancer Manag Res 2020;12:6947-58. [Crossref] [PubMed]
- Tanaka K, Yu HA, Yang S, et al. Targeting Aurora B kinase prevents and overcomes resistance to EGFR inhibitors in lung cancer by enhancing BIM- and PUMA-mediated apoptosis. Cancer Cell 2021;39:1245-1261.e6. [Crossref] [PubMed]
- Huang J, Zhang Q, Shen J, et al. Multi-omics analysis identifies potential mechanisms of AURKB in mediating poor outcome of lung adenocarcinoma. Aging (Albany NY) 2021;13:5946-66. [Crossref] [PubMed]
- Shin J, Kim TW, Kim H, et al. Aurkb/PP1-mediated resetting of Oct4 during the cell cycle determines the identity of embryonic stem cells. Elife 2016;5:e10877. [Crossref] [PubMed]
- Zhang J, Lin X, Wu L, et al. Aurora B induces epithelial-mesenchymal transition by stabilizing Snail1 to promote basal-like breast cancer metastasis. Oncogene 2020;39:2550-67. [Crossref] [PubMed]
- Bonet C, Giuliano S, Ohanna M, et al. Aurora B is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem 2012;287:29887-98. [Crossref] [PubMed]
- Madejón A, Sheldon J, Francisco-Recuero I, et al. Hepatitis C virus-mediated Aurora B kinase inhibition modulates inflammatory pathway and viral infectivity. J Hepatol 2015;63:312-9. [Crossref] [PubMed]
- He SJ, Shu LP, Zhou ZW, et al. Inhibition of Aurora kinases induces apoptosis and autophagy via AURKB/p70S6K/RPL15 axis in human leukemia cells. Cancer Lett 2016;382:215-30. [Crossref] [PubMed]
- Moreira-Nunes CA, Mesquita FP, Portilho AJS, et al. Targeting aurora kinases as a potential prognostic and therapeutical biomarkers in pediatric acute lymphoblastic leukaemia. Sci Rep 2020;10:21272. [Crossref] [PubMed]
- Ramani P, Sowa-Avugrah E, May MT. High proliferation index, as determined by immunohistochemical expression of Aurora kinase B and geminin, indicates poor prognosis in neuroblastomas. Virchows Arch 2015;467:319-27. [Crossref] [PubMed]
- Xiao J, Zhang Y. AURKB as a Promising Prognostic Biomarker in Hepatocellular Carcinoma. Evol Bioinform Online 2021;17:11769343211057589. [Crossref] [PubMed]
- Tovuu LO, Utsunomiya T, Imura S, et al. The role of Aurora B expression in non-tumor liver tissues of patients with hepatocellular carcinoma. Int J Clin Oncol 2014;19:622-8. [Crossref] [PubMed]
- Yasen M, Mizushima H, Mogushi K, et al. Expression of Aurora B and alternative variant forms in hepatocellular carcinoma and adjacent tissue. Cancer Sci 2009;100:472-80. [Crossref] [PubMed]
- Lin ZZ, Jeng YM, Hu FC, et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer 2010;10:461. [Crossref] [PubMed]
- Tanaka S, Arii S, Yasen M, et al. Aurora kinase B is a predictive factor for the aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br J Surg 2008;95:611-9. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [Crossref] [PubMed]
- Gao X, Jiang A, Shen Y, et al. Expression and clinical significance of AURKB gene in lung adenocarcinoma: Analysis based on the data-mining of bioinformatic database. Medicine (Baltimore) 2021;100:e26439. [Crossref] [PubMed]
- Nie M, Wang Y, Yu Z, et al. AURKB promotes gastric cancer progression via activation of CCND1 expression. Aging (Albany NY) 2020;12:1304-21. [Crossref] [PubMed]
- Xu X, Zhang HL, Liu QP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol 2019;70:1133-44. [Crossref] [PubMed]
- Erstad DJ, Tanabe KK. Prognostic and Therapeutic Implications of Microvascular Invasion in Hepatocellular Carcinoma. Ann Surg Oncol 2019;26:1474-93. [Crossref] [PubMed]
- Lei Z, Li J, Wu D, et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus-Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg 2016;151:356-63. [Crossref] [PubMed]
- Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025-41. [Crossref] [PubMed]
- Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507-19. [Crossref] [PubMed]


