
Association of CYP2D6 genotype predicted phenotypes with oxycodone requirements and side effects in children undergoing surgery
Introduction
Children undergo 3.9 million surgeries every year in the United States. Safe and effective postoperative analgesia is critical to successful recovery after invasive surgery in children (1,2). Despite efforts to minimize opioid use for postoperative analgesia, opioids still play a major role in postoperative pain control. Postoperative pain management remains sub-optimal as 40% of patients suffer moderate-to-severe acute pain after surgery, and 40% experience opioid side-effects (3,4). In recent times, there has been a shift in analgesic utilization towards oxycodone (5). Annual prescription prevalence for oxycodone is 2,116 per 100,000 patients, making it the most widely prescribed oral opioid in children in the United States (6), despite its high interindividual response variability and higher risk for opioid dependence (7). Superior oral bioavailability, increased central nervous system permeability, controlled-release formulation and advertised favorable side-effect profile compared to morphine have contributed to oxycodone’s commercial success (8-10).
Lalovic et al., demonstrated that 45%±21% of oxycodone is N-demethylated by cytochrome P450 enzymes (CYP3A4/5) to noroxycodone, noroxymorphone, alpha- and beta-noroxycodol; 11%±6% of oxycodone is O-demethylated via CYP2D6 to oxymorphone, alpha- and beta-oxymorphol; 8%±6% of oxycodone is 6-keto-reuced to alpha- and beta-oxycodol (11,12) (Figure 1). Since oxycodone’s metabolism is mostly dependent on the cytochrome P450 (CYP) family of liver enzymes including CYP2D6 and CYP3A4/5 (9,11), patients with CYP allelic variations could present with unpredictable level of analgesia and side-effects. CYP2D6 is a known polymorphic gene with >130 alleles of defined phenotypes [poor metabolizers (PM), intermediate metabolizers (IM), normal metabolizers (NM) or ultra-rapid metabolizers (UM)]. Different phenotypes are the result of enzyme activity conferred by allelic variations (13). Altered activity CYP2D6 phenotypes have been implicated in poor analgesic responses, increased side effects and even mortality from oral opioids such as codeine (14,15).
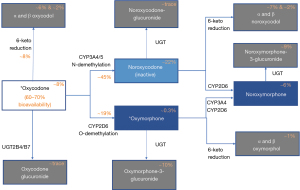
Unlike codeine which is a prodrug and only its CYP2D6 metabolite is active, oxycodone is an active parent drug. However, it is metabolized by CYP2D6 to oxymorphone which has a 10–60‐fold higher µ‐opioid receptor affinity (16), and similar abuse liability (17). Oxymorphone/oxycodone ratios were shown in pharmacokinetic (PK) studies to be lower in CYP2D6 PM compared to NM/IM (18). Despite the knowledge of CYP2D6 variations and their influences on oxycodone exposure (19,20), the impact of the polymorphic CYP2D6 on oxycodone clinical effects is not clear (21). Although a handful of studies show CYP2D6 phenotypes impact oxycodone analgesia (22) and serious adverse effects including mortality (23,24), studies in pediatric populations show conflicting findings (19,22). Per recent Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines, “there is insufficient evidence and confidence to provide a recommendation to guide clinical practice at this time for oxycodone” based on CYP2D6 genotype (19). However, Ramsey et al. suggested possible merit in genotype-guided prescription of drugs metabolized by CYP2D6, including oxycodone due to its high annual use amongst United States’ pediatric patients (2,116 per 100,000 patients; 95% CI: 2,097–2,135) (6).
In this study, we aimed to determine associations between CYP2D6 genotype-predicted phenotypes with oxycodone requirements for analgesia, risk of respiratory depression (RD) and emesis in post-surgical children. We hypothesized that compared to NM, PM and UM will have different oxycodone requirements to maintain analgesia. Also, RD and emesis would be associated with phenotype in high oxycodone use groups but not in low oxycodone use groups. In addition, we explored whether genotype availability and provider guidance would influence oral opioid prescription patterns. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-2022-58/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of Cincinnati Childrens (Protocol# 2013-0853) and informed consent was obtained from all individual participants for the prospective part of the study (2012–2015) and was waived for the retrospective data review (2016–2018). Retrospective chart review of data from two cohorts undergoing Nuss procedure or posterior spine fusion (for idiopathic scoliosis) between 2012 and 2018 was conducted. The first cohort included patients recruited for a prospective preemptive CYP2D6 genotyping study between 2012–2015. The second cohort consisted of patients who were genotyped for CYP2D6 preoperatively as part of clinical management between 2016–2018. Patients in both cohorts had CYP2D6 genotyping completed and resulted before surgery. Inclusion criteria for chart review: American Society of Anesthesiologists classification 1–3, diagnosis of pectus excavatum or idiopathic scoliosis undergoing Nuss procedure or spine fusion respectively, with CYP2D6 genotyping results available before or at the time of surgery. Exclusion criteria included presence of obesity (BMI >30 kg/m2), developmental delays, age <8 years, liver/renal disease (based on patient’s preoperative medical history), history of substance abuse/opioid use preoperatively.
All patients underwent surgery using standardized anesthesia protocols (general endotracheal anesthesia using total intravenous anesthesia/intraoperative neuromonitoring for spine fusion and inhalation anesthesia/muscle relaxant for Nuss procedure). Postoperatively, spine fusion patients received patient-controlled analgesia (morphine or hydromorphone) and were transitioned to oral opioids on postoperative days (POD) one or two. All patients received ondansetron intravenously every 6 hours for emesis prophylaxis. Pectus patients received a thoracic epidural which was continued until POD 2 (2016–2018) and POD 3 (2012–2015). All patients were monitored using continuous electrocardiography (heart rate), thoracic Impedance plethysmography (respiratory rate), pulse oximetry (oxygen saturation) and intermittent blood pressure measurement on the floor after surgery. All subjects were offered oral opioids every four hours (either oxycodone 0.1–0.2 mg/kg or hydromorphone 0.04–0.08 mg/kg). Opioid CYP2D6 Panel result approved by institutional Genetic Pharmacology Service was available in electronic medical records (EMR) (Epic Systems, Verona, WI, USA) for clinical use. An EPIC alert notified the clinician that a pharmacogenetic (PGx) test result was available for the medication. Before 2016 (during the preemptive genotyping study), pain physicians were provided education about the genotyping alerts on EPIC, but not directed to use the genotyping results available to them while prescribing oxycodone after surgery. Between 2016–2018, patients were clinically genotyped and postoperative pain team was directed to administer CYP2D6 non-dependent oral opioid (hydromorphone/morphine instead of oxycodone for CYP2D6 UM and PM) per the genotype recommendation on genotyping result. For the exploratory aim, patients undergoing surgery between 2012–2015 (cohort 1) were considered the control group and those between 2016–2018 (cohort 2) were the intervention group.
Data collection
Patients for the chart review were identified based on prospectively recruited subjects (2012–2015) and list of patients who underwent pectus surgery (2016–2018), and satisfied criteria. The patient list was provided to EPIC data informatics team to retrieve the data described below. The data was manually verified and missing data retrieved by authors (SM, JM). Data variables and outcomes assessments were focused on POD after discontinuation of regional analgesia for assessing oxycodone effects (POD 2: cohort 2 and POD3: cohort 1).
Patient characteristics: demographics (age, sex, race, weight).
Surgical data: surgery type, date of surgery, diagnosis.
Oral opioid and doses: names of any oral and intravenous opioid administered (oxycodone, hydromorphone, morphine), times for administration and opioid doses were collected.
CYP2D6 inhibitor use: concomitant use of fluoxetine, bupropion, paroxetine, quinidine, terbinafine and duloxetine was documented.
Pain assessment: pain scores [numerical rating scale (25) (NRS: 0–10)] assessed by nursing and documented in EMR were obtained. Pain scores after epidural removal or PCA discontinuation and after oral opioid was started were included for evaluation.
RD: documentation of at least one occurrence of: (I) needing supplemental oxygen (including blow by) or (II) respiratory rate ≤10 for age 6–10 years; RR ≤8–10 for older than 10 years or (III) SpO2 <90% while spontaneously breathing room air.
Vomiting was scored as a binary variable (yes or no) by reviewing output on the EMR.
Other relevant medications: EMR was evaluated for administration of known strong CYP2D6 inhibitors (Fluoxetine, Bupropion, Paroxetine, Quinidine, Terbinafine and Duloxetine) and recorded.
Genotyping
DNA was collected from blood/saliva for CYP2D6 genotyping several days before surgery in all subjects. In both cohorts, genotyping results were available before day of surgery. The Cincinnati Children’s Hospital Medical Center (CCHMC) CLIA approved, CAP certified molecular genetics laboratory performed CYP2D6 genotyping using the TaqMan allelic discrimination system (Applied Biosystem, Forest City, CA, USA) on a low density microarray for all alleles except CYP2D6*5 allele (full gene deletion) and CYP2D6 duplication (Table 1). Allele designations was performed by the most current and validated Autocaller (1.1) and laboratory designed allele calling tools. A known genotype control was run with each batch to ensure the quality of the allelic calls. CYP2D6*5 allele (full gene deletion) and CYP2D6 duplication were detected by long-PCR according to the method described by Løvlie et al. (26) These assays allow distinction between the CYP2D6 gene and the non-functional pseudogenes CYP2D7 and CYP2D8 but do not detect allele-specific copy number. Turnaround time for results was 2 business days.
Table 1
| Enzyme activity | CYP2D6 allele (variant reference number) |
|---|---|
| Full | *1 (N/A), *2 (rs16947), *2A (hCV32407252), *35 (rs769258) |
| Reduced | *9 (rs5030656), *10 (rs1065852), *17 (hCV2222771), *41 (hCV34816116) |
| None | *3 (hCV32407232), *4 (rs3892097), *5 (deleted), *6 (rs5030655), *7 (rs5030867), *8 (rs5030865), *11 (rs5030863), *14 (rs5030865), *15 (hCV32407245), *18 (hCV32407220), *19 (hCV32407233), *20 (hCV72649949), *40 (hCV32407240), *42 (hCV72649935), *44 (hCV32407228) |
CYP2D6*5 allele (full gene deletion) and CYP2D6 duplication were detected by long-PCR; *, empirical assignment of detected variants into star (*) alleles defined by the Pharmacogene Variation Consortium (https://www.pharmvar.org/gene/CYP2D6). PCR, polymerase chain reaction.
Phenotyping
Based on CYP2D6 alleles, allelic activity scores and total activity scores (TAS) were determined, and phenotypes (PM, IM, NM and UM) defined, according to latest guidelines (Table 2) (27). In subjects using CYP2D6 strong/moderate inhibitors, we accounted for phenoconversion by multiplying the TAS by inhibitor factor: 0 for strong (e.g., bupropion, fluoxetine) and 0.5 for moderate (e.g., duloxetine) inhibitors, as has been done previously (28,29). For the exploratory aim, we used EMR documented phenotypes (which were based on previous CPIC guidelines) (30).
Table 2
| CYP2D6 phenotype | Activity score | Examples of CYP2D6 diplotypes |
|---|---|---|
| Ultrarapid metabolizer | >2.25 | *1/*1xN, *1/*2xN, *2/*2xN, *1x2/*9 |
| Normal metabolizer | 1.25≤x≤2.25 | *1/*10, *1/*41, *1/*9, *1/*1, *1/*2, *2x2/*10 |
| Intermediate metabolizer | 0<x<1.25 | *4/*10, *4/*41, *10/*10, *10/*41, *41/*41, *1/*5 |
| Poor metabolizer | 0 | *3/*4, *4/*4, *5/*5, *5/*6 |
*, indicates star alleles based on empirical assignment of detected variants defined by the Pharmacogene Variation Consortium (https://www.pharmvar.org/gene/CYP2D6).
Outcomes
Primary outcome was oxycodone analgesia which was measured as oxycodone requirements in morphine equivalents (MEq)/kg relative to total MEq/kg use, and secondary outcomes were risk for RD and emesis.
Statistical analysis
Descriptive statistics (mean and standard deviation, or median and interquartile range for continuous variables, and frequencies and percentages for categorical variables) were generated for demographics and other data characteristics, and normality for continuous variables was assessed. Pain experienced over time in the immediate postoperative period is captured well by the area under curve (AUC) which was calculated using trapezoidal rule for area under the curve of pain scores over time period [as in prior studies (2,31)]. AUC% for pain scores <4 (mild pain), were also calculated as %AUC corresponding to pain score <4/10. Although pain AUC was initially the analgesia outcome measure, considering the opioid drug and dose changes that were made clinically to maintain analgesia, relative use of oxycodone (as the CYP2D6 dependent opioid) was analyzed post-hoc as primary outcome measure. MEq/kg were calculated for all oral and intravenous opioids. Oxycodone (CYP2D6 dependent) and CYP2D6 non-dependent (morphine, hydromorphone) opioid MEq/kg were calculated separately and ratio of oxycodone requirements relative to total MEq/kg obtained. We first determined univariate associations of outcomes with categorical independent variables (sex, race, surgery type, need for opioid change), either two-sample t-tests (if normal) or Wilcoxon rank-sum test (if not normal) and continuous independent variables (age, TAS) using Pearson correlation (if both normal), or Spearman correlation (if any nonnormal). We checked for correlation of covariates with CYP2D6 phenotype. Multivariable linear regression model was used to investigate association of CYP2D6 genotype predicted phenotype (PM, IM, UM vs. NM) with primary. Stratified logistic regression was conducted in low (oxycodone/total MEq <0.5) and high (and oxycodone/total MEq >0.5) oxycodone use groups, with application of firth correction due to quasi-complete separations. Covariates were included in multiple regression models if associated with outcome (P<0.1 on univariate analyses) but not correlated with CYP2D6 phenotype. For the exploratory aim, we compared control (cohort 1) and EMR directed groups (cohort 2) for first oral opioid medication prescribed (CYP2D6 vs. non-CYP2D6 substrate) using Fisher’s exact tests. Breslow-Day test was used to evaluate odds ratios between the groups. P<0.05 was used for denoting statistical significance.
Results
The combined cohort comprised of 193 subjects in all. Average age was 15.9 years (SD 0.25), 55/193 (28.5%) females, 181/193 (93.78%) white, weight 55.26 kg (SD 13.45); 77.72% underwent pectus surgery. The cohort overall had an average CYP2D6 TAS of 1.435 (SD 0.647) and included 101 NM, 76 IM, 10 PM and 6 UM phenotypes (two IM phenotypes were on CYP2D6 inhibitors and were phenoconverted to PM). Oxycodone was the most commonly prescribed first oral opioid (N=184; 9 hydromorphone). In 32 subjects, oxycodone was changed to hydromorphone while hydromorphone was switched to oxycodone in 32 subjects. Mean (SD) of AUC of pain for the cohort was 85.98 (3.58). Outcomes and cohort characteristics by CYP2D6 phenotypes are provided in Table 3.
Table 3
| Independent variables | CYP2D6 phenotype | |||
|---|---|---|---|---|
| PM (N=10) | IM (N=76) | NM (N=101) | UM (N=6) | |
| Sex (n, %) | ||||
| Female | 1 (1.82) | 18 (32.73) | 34 (61.82) | 2 (3.64) |
| Male | 9 (6.52) | 58 (42.03) | 67 (48.55) | 4 (2.90) |
| Race (n, %) | ||||
| Non-white | 1 (8.33) | 6 (50.00) | 5 (41.67) | 0 (0.00) |
| White | 9 (4.97) | 70 (38.67) | 96 (53.04) | 6 (3.31) |
| Surgery type (n, %) | ||||
| Pectus | 10 (6.67) | 63 (42.00) | 74 (49.33) | 3 (2.00) |
| Spine | 0 (0.00) | 13 (30.23) | 27 (62.79) | 3 (6.98) |
| Respiratory depression, yes (n, %) | 6 (13.95) | 11 (25.58) | 26 (60.47) | 0 (0.00) |
| Emesis, yes (n, %) | 3 (5.88) | 20 (39.22) | 25 (49.02) | 3 (5.88) |
| Need for oral opioid change, yes (n, %) | 4 (8.16) | 18 (36.73) | 25 (51.02) | 2 (4.08) |
| %AUC above pain score 4/10, yes (n, %) | 5 (3.68) | 56 (41.18) | 70 (51.47) | 5 (3.68) |
| Age (years), median (IQR) | 15.4 (14.5, 20.2) | 15.7 (14.4, 16.8) | 15.4 (14.2, 16.8) | 15.4 (13.9, 20.7) |
| Total activity score, median (IQR) | 0.0 (0.0, 0.0) | 1.0 (1.0, 1.0) | 2.0 (1.5, 2.0) | 3.0 (3.0, 3.0) |
| Pain area under curve, median (IQR) | 50.2 (43.5, 74.3) | 93.5 (45.8, 127.7) | 77.8 (48.0, 119.5) | 111.0 (79.7, 138.2) |
| Oxycodone MEq/total MEq, median (IQR) | 0.6 (0.0, 0.7) | 0.8 (0.7, 0.9) | 0.9 (0.7, 1.0) | 0.2 90.0, 0.9) |
PM, poor metabolizer; IM, intermediate metabolizer; NM, normal metabolizer; UM, ultrarapid metabolizer; AUC, area under curve for pain scores over time; IQR, interquartile range; MEq, morphine equivalents.
Pain AUC
On comparing AUC by age, surgery, sex and opioid choice, we found female sex and spine surgery had higher AUC, but no differences by opioid choice or age. Median (IQR) of AUC was 111.22 (76.07, 138.17) in females vs. 71.10 (41.15, 115.37) in males; P<0.001); spine surgery [median (IQR) 105.59 (73.89, 137.28) compared to 75.4 (41.7, 117.5) in pectus; P=0.008]. oxycodone use [median (IQR) of 82.62 (46.28, 125.95) vs. hydromorphone use 78.74 (61.5, 103.76); P=0.773] and age (Spearman correlation coefficient 0.102, P=0.179). In those subjects receiving oxycodone only, average pain AUC was 123.2 (SD 52.8) for UM, 88.9 (SD 51.9) for IM, 84.8 (SD 44.3) for NM and 67.9 (39.9) for PM. CYP2D6 phenotype was not found to be a significant predictor for pain AUC (P=0.643).
Oxycodone requirements
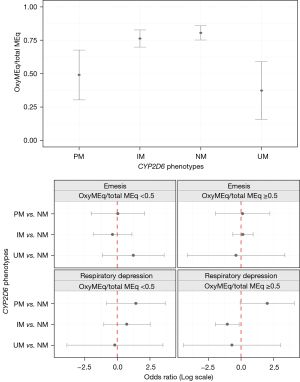
Of the independent variables evaluated for association with oxycodone/total MEq, surgery type was significant. Oxycodone was used more often in spine surgery compared to pectus surgery [1.0 (95% CI: 0.805–1.000) vs. 0.789 (95% CI: 0.636–1.000), P=0.013]. However, surgery type was not included in multiple regression due to being correlated to CYP2D6 phenotype (P=0.041). Spearman correlation for AUC with oxycodone MEq/total MEq was −0.133 (P=0.079). Multivariate regression results for CYP2D6 phenotype, adjusted for pain AUC are given in Table 4, with CPY2D6 phenotype being a significant predictor. Compared to NM, least squares means of oxycodone MEq/total MEq were lower for PM [−0.301 (95% CI: −0.496, −0.106), P=0.008] and UM [−0.443 (95% CI: −0.666, −0.221), P<0.001], but not statistically significant for IM vs. NM [−0.035 (95% CI: −0.120, 0.049), P=1.00]. Predicted oxycodone MEq/total MEq plotted against CYP2D6 phenotypes are presented in Figure 2. Phenotype was a significant predictor (P<0.001) with both PM and UM phenotypes using lower oxycodone/total MEq doses compared to NM [−0.316 (SE 0.098), P=0.005 and −0.432 (SE 0.113), P<0.001 respectively].
Table 4
| Outcomes | Predictors | Category | Least squares means | P value | |||
|---|---|---|---|---|---|---|---|
| Estimate | SE | Lower CL | Upper CL | ||||
| Oxycodone/total MEq | CYP2D6 phenotype | IM | 0.781 | 0.036 | 0.709 | 0.853 | <0.001 |
| PM | 0.516 | 0.097 | 0.324 | 0.707 | |||
| UM | 0.373 | 0.109 | 0.158 | 0.588 | |||
| NM | 0.816 | 0.030 | 0.758 | 0.875 | |||
| Surgery | Spine | 0.651 | 0.053 | 0.546 | 0.757 | 0.233 | |
| Pectus | 0.592 | 0.039 | 0.514 | 0.669 | |||
MEq, morphine equivalents; SE, standard error; CL, confidence level; IM, intermediate metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer; NM, normal metabolizer.
Risk of RD
Univariate analyses identified no variables associated with RD (Table 3). Stratified regression showed that CYP2D6 phenotypes were not associated with RD when oxycodone/total MEq <0.5 (P=0.614) but was associated in the oxycodone/total MEq >0.5 group (P=0.018) (Table 5). IM had lower odds for RD compared to NM (0.346; 95% CI: 0.140−0.855; P=0.021). While UM had lower odds and PM higher odds for RD compared to NM, they were not statistically significant. Predicted odds for risk of RD for different phenotypes (PM, IM and UM) vs. NM are presented in Figure 2.
Table 5
| Side effects | OxyMEq/total MEq | CYP2D6 phenotype | OR (lower CL, upper CL) | P value |
|---|---|---|---|---|
| Respiratory depression | <0.5 (n=40) | IM vs. NM | 2.088 (0.354, 12.309) | 0.614 |
| PM vs. NM | 4.142 (0.435, 39.412) | |||
| UM vs. NM | 0.829 (0.021, 33.202) | |||
| ≥0.5 (n=151) | IM vs. NM | 0.346 (0.140, 0.855) | 0.018 | |
| PM vs. NM | 7.408 (0.927, 59.214) | |||
| UM vs. NM | 0.494 (0.012, 20.974) | |||
| Emesis | <0.5 (n=40) | IM vs. NM | 0.699 (0.161, 3.038) | 0.634 |
| PM vs. NM | 1.044 (0.134, 8.149) | |||
| UM vs. NM | 3.409 (0.318, 36.601) | |||
| ≥0.5 (n=151) | IM vs. NM | 1.144 (0.529, 2.477) | 0.981 | |
| PM vs. NM | 1.120 (0.139, 9.027) | |||
| UM vs. NM | 0.671 (0.016, 28.667) |
OxyMEq, oxycodone morphine equivalents; OR, odds ratio; CL, confidence level; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
Risk of emesis
Pectus surgery was associated with higher emesis (30.94%) compared to spine surgery (7.5%) (P=0.003) but was not included in multiple regression due to being correlated to CYP2D6 phenotype (P=0.041). Stratified regression (Table 5) did not find CYP2D6 phenotypes to be associated with risk of emesis. Predicted odds for risk of emesis for different phenotypes (PM, IM and UM) vs. NM are presented in Figure 2.
Genotype guidance exploratory aim
There were 66 subjects in control (2012–2015 non directed genotype group) and 127 subjects included the genotype directed group (2016–2018). Characteristics of the groups are provided in Table 6. In the control group, there was no significant difference in prescribing of CYP2D6 dependent opioids [91.24% of NM/IM vs. 66.67% of PM/UM (P=0.129)]; in the genotype-directed group, there was a significant difference [94.64% of NM/IM vs. 28.57% of PM/UM were prescribed CYP2D6 dependent opioids (P<0.001)]. The Odds for prescribing non-CYP2D6 opioids to PM/UM compared to NM/IM was 5.2 in the control group and 44.17 in the genotype-directed group (Breslow-Day test, P=0.105). Logistic regression showed that PM/UM phenotypes in genotype directed group had a lower chance of being prescribed CYP2D6 opioids (effect size =−2.775; SE: 1.566; P=0.076).
Table 6
| Independent variables | Control group (N=66) | Genotype guided (N=127) |
|---|---|---|
| Age (years), mean (SD) | 15.2 (2.6) | 16.2 (3.8) |
| Sex, female (n, %) | 34 (51.5%) | 21 (16.5%) |
| Race, White (n, %) | 62 (93.9%) | 119 (93.7%) |
| Surgery | Pectus 23; spine 43 | Pectus 127 |
| Phenotype distribution | ||
| Poor/ultrarapid metabolizer | 1/5 | 7/1 |
| Intermediate/normal metabolizer | 3/57 | 6/112 |
| TAS, mean (SD) | 1.55 (0.64) | 1.38 (0.59) |
| First opioid prescribed, n | ||
| Oxycodone | 56 | 108 |
| Hydromorphone | 7 | 11 |
| Need for medication/dose changes (n, %) | 17 (26.9%) | 31 (25.8%) |
| Pain AUC, median (IQR) | 97.1 (69.6, 131.3) | 72.6 (39.7, 116.3) |
| Emesis, yes/no (% yes) | 8/53 (13.11%) | 38/80 (32.20%) |
| Respiratory depression, yes/no (% yes) | 14/46 (23.33%) | 24/90 (20.34%) |
SD, standard deviation; TAS, total activity score; AUC, area under curve; IQR, inter quartile range.
Discussion
In this retrospective study, we evaluated whether oxycodone requirements and side effects were associated with CYP2D6 phenotypes. Genotype predicted CYP2D6 phenotype was associated with lower oxycodone requirements to achieve analgesia in PM and UM compared to NM and risk for RD in the high oxycodone dose group only. No associations were identified for emesis with any phenotype. We also demonstrated that genotyping directed guidance to prescribe oral opioids, compared to reliance on passive EMR alerts resulted in a lower chance of being prescribed oxycodone in poor and UM, compared to NM and IM phenotypes.
Balyan et al. and Stamer et al. have demonstrated lower oxymorphone/oxycodone ratios in PM in adults and children (18,21). Since oxymorphone has 10–60-fold increased potency and affinity for mu opioid receptors compared to oxycodone, we expected UM to have lower need for oxycodone to achieve similar analgesia, and this is what we found. However, contrary to expectation, risk for oxycodone side effects were not found to be higher in UM vs. NM. Our findings are aligned with previous reports of higher potency of oxymorphone with comparable/fewer side effects, although this was not previously demonstrated in the context of CYP2D6 phenotypes (32).
To our surprise, we also found lower oxycodone requirements to maintain analgesia in PM compared to NM. This might be due to increased effects of an already active drug. In addition, oxycodone has superior blood-brain barrier (BBB) penetration with concentrations reaching 3- to 6-fold higher in the brain than in the plasma (10,33,34). While UM phenotype was not associated with RD or emesis, IM was associated with lower risk for RD in high dose oxycodone group, while PM had nominally higher risk. In NM, the oxycodone/oxymorphone plasma and brain concentration ratios are 43:1 and 78:1 respectively, and this increases to 110:1 and 540:1 in PM, and decreases to 32:1 and 57:1 in UM (35). Although previous studies have shown oxycodone to be the major contributor of analgesia after oral and intravenous administration, 83.02% and 94.76% respectively; while oxymorphone only plays a minor role (15.77% and 4.52% after oral and intravenous administration, respectively), our findings suggest both higher oxycodone effects in PM and oxymorphone in UM may have clinically significant effects (36,37). Thus, while the Royal Dutch Pharmacists Association-Pharmacogenetics Working Group (DPWG) have concluded that no change in oxycodone prescribing is required based on CYP2D6 genotype (38), our study demonstrates that there might be potential differences in phenotype effects, which need further investigation in bigger samples in a prospective randomized controlled study.
However, the effect sizes for phenotypes on outcomes are small. This might be because in order for patients to experience clinically significant disturbance in oxycodone metabolism, both CYP2D6 and CYP 3A4/5 pathways may need to be compromised. A “two-hit phenomenon” is supported by Gronlund and colleagues (39) who illustrated that with concomitant administration of CYP2D6 and CYP3A4 inhibitors, the AUC of oxycodone concentration increased 2.9-fold with subsequent increases in pain scores, and drowsiness.
Contrary to our findings, Zwisler et al. found no difference in total oxycodone consumption between CYP2D6 PM in a genotype blinded study in post-surgical patients (40). However, the same authors found that antinociceptive effects of oxycodone depended on oxymorphone in experimental pain models with higher pain thresholds after nerve stimulation and lower AUC after cold pressor tests in EM compared to PM (41). It is possible that experimental and clinical pain phenotypes may be differently influenced by CYP2D6 phenotypes. Furthermore, the findings of our exploratory aim are aligned with the report by Thomas and colleagues who demonstrated that genotype-guided care influenced opioid prescription of oxycodone. In their high risk group (IM/PM and UM), 72% were prescribed an alternative opioid vs. 0% in their usual care participants (42).
Limitations of this study include under representation of UM patients in our sample population. As summarized by Gaedigk et al., prevalence of CYP2D6 phenotypes differ between different race/ethnic populations (13). For example, UM phenotypes are highest in Oceanian (21.2%) followed by Ashkenazi Jewish (11.5%) and Middle Eastern (11.2%) populations. PM phenotypes are highest in Europeans (5.4%) and in the Ashkenazi Jewish population (6%). Our cohort was primarily white and was unable to capture UM phenotypes adequately (14). Given the discrepancy between findings of associations of IM with outcomes, which are difficult to explain, we cannot rule out findings as a result of chance alone. Larger prospective studies are needed to validate the findings of this study. Future studies may also consider methods to preferentially include genotype predicted PM/UMs. Additionally, all patients received similar antiemetics (ondansetron) also metabolized by CYP2D6. Therefore, it is possible that in PM phenotypes, ondansetron may have a superior effectiveness and dampen the incidence of emesis and hence skew our results for oxycodone associated emesis (43). Lastly, other genetic variants can affect oxycodone outcomes (for example, mu opioid receptor variant A118G and ATP binding cassette variants C3435T and G2677T/A), which were not studied here (44). Future studies may also need to evaluate the level of CYP2D6 DNA methylation since its expression is epigenetically regulated (45).
One confounder for enteric oxycodone is the wide interindividual variation in absorption. In a study by Kokki and colleagues (46) a large variation in the rate and extent of absorption of buccal and enteric oxycodone in children ages 6–93 months was observed. Zwisler et al. illustrated that one hour after taking oral oxycodone, plasma concentrations in healthy subjects varied considerably (2.99–104.25 ng/mL in PM and 0.12–72.68 ng/mL in EM) (41). Thus, it is ideal to measure plasma oxycodone concentrations and exposure. Another confounder is the phenomenon of phenoconversion, whereby IMs behave as PM phenotype. This may occur when there is co-administration of oxycodone and CYP2D6 inhibitors, which has been shown to influence pain management (47). Although the magnitude of phenoconversion in our cohort was small, only 2 subjects were impacted—phenoconverted from IM to PM, drugs used to treat pediatric behavioral disorders are rampant and several of these are CYP2D6 inhibitors. Hence, this may be an important area for future investigations and generalizability to other surgical populations in a more diverse cohort of children.
Conclusions
In this study we investigated association between CYP2D6 predicted phenotypes, oxycodone requirements and side-effects. We found relative oxycodone requirements to maintain analgesia were lower in PM and UM compared to NM, leading us to posit these might be due to higher oxycodone and oxymorphone effects in these respective groups. Risk for RD was associated with CYP2D6 phenotype in high oxycodone but not the low oxycodone use groups. Our findings suggest that at extreme phenotypes, increased parent oxycodone and oxymorphone may influence clinical outcomes but oxymorphone may have a better safety profile. We also found that EMR available CYP2D6 phenotype information significantly influenced oral opioid prescription pattern.
Acknowledgments
We thank Qing Duan, PhD, staff statistician from the Department of Biostatistics and Epidemiology, Cincinnati Children’s Hospital Medical Center for her contributions to statistical analyses for the revised manuscript.
Funding: This research was part of a single site eMERGE II project initiated and funded by the NHGRI through grants U01HG006828 and U01HG008666. This work was supported by the US Department of Health & Human Services National Institutes of Health (NIAMS) under grant Award R01AR075857 to Chidambaran (PI), Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Numbers, R01HD089458 (PI: Sadhasivam), R21HD094311(PI: Sadhasivam) and R01HD096800 (PI: Sadhasivam). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Authors were supported by departmental salaries.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-2022-58/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-2022-58/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-2022-58/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-2022-58/coif). SM declares single site eMERGE II project initiated and funded by the NHGRI through grants U01HG006828 and U01HG004438 (Prows); research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Numbers, R01HD089458 (PI: Sadhasivam), R21HD094311(PI: Sadhasivam) and R01HD096800 (PI: Sadhasivam). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. CAP received funding from National Human Genome Research Institute at National Institutes of Health, USA, through grants U01HG006828 and U01HG004438, Elsevier Inc. ($250, chapter in Wong’s Nursing Care of Infants and Children, 12th edition), and Patent US 10,662,476 B2—Personalized pain management and anesthesia: preemptive risk identification and therapeutic decision support. SS received funding from R01HD089458 (PI: Sadhasivam), R21HD094311(PI: Sadhasivam), R01HD096800 (PI: Sadhasivam), R01HD089458 (PI: Sadhasivam), R21HD094311(PI: Sadhasivam) and R01HD096800 (PI: Sadhasivam). And received pay from UpToDate: Anesthesia for Tonsillectomy and NeurOptics: Morphine in Tonsillectomy. SS is one of the inventors in the approved U.S. patents focused on opioid pharmacogenetics: U.S. Patent No. 9944985, 10662476, 16/850537, 16/946401, 16/946399, 10878939. He is the founder and chief medical officer of OpalGenix, Inc. There is no financial conflict with the current article. 9944985, 10662476, 16/946401, 16/850537 also include Dr. Chidambaran and Cindy Prows. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of Cincinnati Childrens (Protocol# 2013-0853) and informed consent was obtained from all individual participants for the prospective part of the study (2012–2015) and was waived for the retrospective data review (2016–2018).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rabbitts JA, Groenewald CB. Epidemiology of Pediatric Surgery in the United States. Paediatr Anaesth 2020;30:1083-90. [Crossref] [PubMed]
- Chidambaran V, Ding L, Moore DL, et al. Predicting the pain continuum after adolescent idiopathic scoliosis surgery: A prospective cohort study. Eur J Pain 2017;21:1252-65. [Crossref] [PubMed]
- Gerbershagen HJ, Pogatzki-Zahn E, Aduckathil S, et al. Procedure-specific risk factor analysis for the development of severe postoperative pain. Anesthesiology 2014;120:1237-45. [Crossref] [PubMed]
- Kozlowski LJ, Kost-Byerly S, Colantuoni E, et al. Pain prevalence, intensity, assessment and management in a hospitalized pediatric population. Pain Manag Nurs 2014;15:22-35. [Crossref] [PubMed]
- Dart RC, Surratt HL, Cicero TJ, et al. Trends in opioid analgesic abuse and mortality in the United States. N Engl J Med 2015;372:241-8. [Crossref] [PubMed]
- Ramsey LB, Ong HH, Schildcrout JS, et al. Prescribing Prevalence of Medications With Potential Genotype-Guided Dosing in Pediatric Patients. JAMA Netw Open 2020;3:e2029411. [Crossref] [PubMed]
- Balyan R, Hahn D, Huang H, et al. Pharmacokinetic and pharmacodynamic considerations in developing a response to the opioid epidemic. Expert Opin Drug Metab Toxicol 2020;16:125-41. [Crossref] [PubMed]
- Oxycodone Kalso E. J Pain Symptom Manage 2005;29:S47-56. [Crossref] [PubMed]
- Kinnunen M, Piirainen P, Kokki H, et al. Updated Clinical Pharmacokinetics and Pharmacodynamics of Oxycodone. Clin Pharmacokinet 2019;58:705-25. [Crossref] [PubMed]
- Boström E, Hammarlund-Udenaes M, Simonsson US. Blood-brain barrier transport helps to explain discrepancies in in vivo potency between oxycodone and morphine. Anesthesiology 2008;108:495-505. [Crossref] [PubMed]
- Lalovic B, Kharasch E, Hoffer C, et al. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin Pharmacol Ther 2006;79:461-79. [Crossref] [PubMed]
- Lalovic B, Phillips B, Risler LL, et al. Quantitative contribution of CYP2D6 and CYP3A to oxycodone metabolism in human liver and intestinal microsomes. Drug Metab Dispos 2004;32:447-54. [Crossref] [PubMed]
- Gaedigk A, Sangkuhl K, Whirl-Carrillo M, et al. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 2017;19:69-76. [Crossref] [PubMed]
- Prows CA, Zhang X, Huth MM, et al. Codeine-related adverse drug reactions in children following tonsillectomy: a prospective study. Laryngoscope 2014;124:1242-50. [Crossref] [PubMed]
- Chidambaran V, Sadhasivam S, Mahmoud M. Codeine and opioid metabolism: implications and alternatives for pediatric pain management. Curr Opin Anaesthesiol 2017;30:349-56. [Crossref] [PubMed]
- Heiskanen T, Olkkola KT, Kalso E. Effects of blocking CYP2D6 on the pharmacokinetics and pharmacodynamics of oxycodone. Clin Pharmacol Ther 1998;64:603-11. [Crossref] [PubMed]
- Babalonis S, Lofwall MR, Nuzzo PA, et al. Pharmacodynamic effects of oral oxymorphone: abuse liability, analgesic profile and direct physiologic effects in humans. Addict Biol 2016;21:146-58. [Crossref] [PubMed]
- Balyan R, Mecoli M, Venkatasubramanian R, et al. CYP2D6 pharmacogenetic and oxycodone pharmacokinetic association study in pediatric surgical patients. Pharmacogenomics 2017;18:337-48. [Crossref] [PubMed]
- Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharmacol Ther 2021;110:888-96. [Crossref] [PubMed]
- Umukoro NN, Aruldhas BW, Rossos R, et al. Pharmacogenomics of oxycodone: a narrative literature review. Pharmacogenomics 2021;22:275-90. [Crossref] [PubMed]
- Stamer UM, Zhang L, Book M, et al. CYP2D6 genotype dependent oxycodone metabolism in postoperative patients. PLoS One 2013;8:e60239. [Crossref] [PubMed]
- Dagostino C, Allegri M, Napolioni V, et al. CYP2D6 genotype can help to predict effectiveness and safety during opioid treatment for chronic low back pain: results from a retrospective study in an Italian cohort. Pharmgenomics Pers Med 2018;11:179-91. [Crossref] [PubMed]
- Boyle KL, Rosenbaum CD. Oxycodone overdose in the pediatric population: case files of the University of Massachusetts Medical Toxicology Fellowship. J Med Toxicol 2014;10:280-5. [Crossref] [PubMed]
- Jannetto PJ, Wong SH, Gock SB, et al. Pharmacogenomics as molecular autopsy for postmortem forensic toxicology: genotyping cytochrome P450 2D6 for oxycodone cases. J Anal Toxicol 2002;26:438-47. [Crossref] [PubMed]
- von Baeyer CL, Spagrud LJ, McCormick JC, et al. Three new datasets supporting use of the Numerical Rating Scale (NRS-11) for children's self-reports of pain intensity. Pain 2009;143:223-7. [Crossref] [PubMed]
- Løvlie R, Daly AK, Molven A, et al. Ultrarapid metabolizers of debrisoquine: characterization and PCR-based detection of alleles with duplication of the CYP2D6 gene. FEBS Lett 1996;392:30-4. [Crossref] [PubMed]
- Caudle KE, Sangkuhl K, Whirl-Carrillo M, et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci 2020;13:116-24. [Crossref] [PubMed]
- Smith DM, Weitzel KW, Elsey AR, et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med 2019;21:1842-50. [Crossref] [PubMed]
- Cicali EJ, Smith DM, Duong BQ, et al. A Scoping Review of the Evidence Behind Cytochrome P450 2D6 Isoenzyme Inhibitor Classifications. Clin Pharmacol Ther 2020;108:116-25. [Crossref] [PubMed]
- Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014;95:376-82. [Crossref] [PubMed]
- Chidambaran V, Zhang X, Martin LJ, et al. DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmgenomics Pers Med 2017;10:157-68. [Crossref] [PubMed]
- Gimbel J, Ahdieh H. The efficacy and safety of oral immediate-release oxymorphone for postsurgical pain. Anesth Analg 2004;99:1472-7. [Crossref] [PubMed]
- Kokki M, Välitalo P, Kuusisto M, et al. Central nervous system penetration of oxycodone after intravenous and epidural administration. Br J Anaesth 2014;112:133-40. [Crossref] [PubMed]
- Boström E, Simonsson US, Hammarlund-Udenaes M. In vivo blood-brain barrier transport of oxycodone in the rat: indications for active influx and implications for pharmacokinetics/pharmacodynamics. Drug Metab Dispos 2006;34:1624-31. [Crossref] [PubMed]
- Deodhar M, Turgeon J, Michaud V. Contribution of CYP2D6 Functional Activity to Oxycodone Efficacy in Pain Management: Genetic Polymorphisms, Phenoconversion, and Tissue-Selective Metabolism. Pharmaceutics 2021;13:1466. [Crossref] [PubMed]
- Kaiko RF, Benziger DP, Fitzmartin RD, et al. Pharmacokinetic-pharmacodynamic relationships of controlled-release oxycodone. Clin Pharmacol Ther 1996;59:52-61. [Crossref] [PubMed]
- Klimas R, Witticke D, El Fallah S, et al. Contribution of oxycodone and its metabolites to the overall analgesic effect after oxycodone administration. Expert Opin Drug Metab Toxicol 2013;9:517-28. [Crossref] [PubMed]
- Matic M, Nijenhuis M, Soree B, et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6 and opioids (codeine, tramadol and oxycodone). Eur J Hum Genet 2022;30:1105-13. [Crossref] [PubMed]
- Grönlund J, Saari TI, Hagelberg NM, et al. Exposure to oral oxycodone is increased by concomitant inhibition of CYP2D6 and 3A4 pathways, but not by inhibition of CYP2D6 alone. Br J Clin Pharmacol 2010;70:78-87. [Crossref] [PubMed]
- Zwisler ST, Enggaard TP, Mikkelsen S, et al. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand 2010;54:232-40. [Crossref] [PubMed]
- Zwisler ST, Enggaard TP, Noehr-Jensen L, et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin Pharmacol Toxicol 2009;104:335-44. [Crossref] [PubMed]
- Thomas CD, Parvataneni HK, Gray CF, et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet Med 2021;23:621-8. [Crossref] [PubMed]
- Edwards A, Teusink-Cross A, Martin LJ, et al. Influence of CYP2D6 metabolizer status on ondansetron efficacy in pediatric patients undergoing hematopoietic stem cell transplantation: A case series. Clin Transl Sci 2022;15:610-8. [Crossref] [PubMed]
- Zwisler ST, Enggaard TP, Noehr-Jensen L, et al. The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam Clin Pharmacol 2010;24:517-24. [Crossref] [PubMed]
- Habano W, Kawamura K, Iizuka N, et al. Analysis of DNA methylation landscape reveals the roles of DNA methylation in the regulation of drug metabolizing enzymes. Clin Epigenetics 2015;7:105. [Crossref] [PubMed]
- Kokki H, Rasanen I, Reinikainen M, et al. Pharmacokinetics of oxycodone after intravenous, buccal, intramuscular and gastric administration in children. Clin Pharmacokinet 2004;43:613-22. [Crossref] [PubMed]
- Cavallari LH, Van Driest SL, Prows CA, et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet Med 2019;21:2255-63. [Crossref] [PubMed]


