
MiR-363 suppresses the tumor growth of natural killer/T-cell lymphoma via the SIRT6/PI3K/AKT axis
Highlight box
Key findings
• MiR-363 is down-regulated in natural killer/T cell lymphoma (NKTCL) and affects proliferation and apoptosis via
What is known and what is new?
• MiR-363 is down-regulated but poorly understood in NKTCL. SIRT6 can function as an oncogene depending on the tumor type and it targets
• This study firstly examines the expression pattern and mechanism of miR-363 in NKTCL tissues and cells. MiR-363 is significantly decreased in NKTCL and suppresses proliferation and apoptosis of SNK-6 cells upon
What is the implication, and what should change now?
• These findings can provide a new target and guidance for the treatment and diagnosis, as well as offer an additional molecular option for future drug development of NKTCL.
Introduction
Natural killer/T-cell lymphoma (NKTCL) is a rare and aggressive tumor type of non-Hodgkin’s lymphoma (1). It appears to have a geographical predilection for Asia (2) and is relatively uncommon in North America and Europe (3). The early treatment of NKTCL usually involves a sequential or simultaneous combination of chemotherapy and radiotherapy (4,5). Clinically, NKTCL is usually accompanied by a poor treatment response and prognosis, and thus, chemotherapy combined with radiotherapy is typically needed to achieve a superior curative effect (6). In spite of the improvement of treatment strategies for NKTCL, the optimal treatment for patients with advanced-stage or recurrent NKTCL is not yet mature and so continues to demonstrate poor results. Therefore, there is an urgent need to explore novel and efficient therapeutic targets.
Micro ribonucleic acids (RNAs) (miRNAs/miRs) are small non-coding RNAs that can regulate gene expression at the post-transcriptional level. Previously, numerous studies have demonstrated the key regulatory role of miRNAs in the pathogenesis of diseases, especially in cancer (7,8). In addition, researchers have also identified a series of abnormally expressed miRNAs, which exert some unknown but vital functions in T-cell differentiation and the development of T-cell malignant tumors (9). To further clarify these unknown roles of miRNAs in NKTCL, researchers established a genome-wide differential miRNAs expression profile of NKTCL, and the findings showed that miRNA-363 was down-regulated in NKTCL (10). Therefore, miRNA-363 might be a molecular biomarker of NKTCL. Yet, the biological role of miRNA-363 in NKTCL progression remains poorly understood although past papers have reported its function in other carcinomas. In previous studies, miR-363 has been confirmed as a tumor-suppressive miRNA that suppresses the growth and metastasis of a variety of malignant tumors, including gastric cancer (11), breast cancer (12), multiple myeloma (13), colorectal cancer (14), and ovarian cancer (15).
As a nuclear protein of the conserved sirtuins family, sirtuin 6 (SIRT6) is also a nicotinamide adenine dinucleotide (NAD)+-dependent deacetylase, which is mainly involved in metabolic homeostasis and stress resistance (16). SIRT6 has been shown to have a double-sided identity in cancer; it functions as a suppressor or oncogene depending on the tumor type (17,18). For example, the up-regulation of SIRT6 could predict poor outcomes and promote NSCLC metastasis (19); however, it is also an important tumor suppressor of pancreatic ductal adenocarcinoma (PDAC), and thus, is regarded as a potential therapeutic signal in the molecular-defined PDAC subgroup (20). In addition, in hepatocellular and prostate carcinomas, the suppression of SIRT6 further emphasizes its sensitivity to chemotherapy and induction of apoptosis (21,22). Previous reports have shown that miRNA regulation alters SIRT6 expression and participates in the progression of a variety of diseases (23,24). These reports highlight the contribution of SIRT6 in tumor development, but its role in NKTCL has not been studied.
It is reported that SIRT6 downregulation in diffuse large B-cell lymphoma (DLBCL) cells suggested enhanced sensitivity to chemotherapy, during which the targets of the phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) pathway are reduced (25). In other words, the PI3K/AKT pathway naturally displays a positive promotion of cell survival (26). Previous literature has shown that the inhibition of PI3K/AKT signaling can promote the apoptosis of primary exudative lymphoma cells (27). Also, significant activation of PI3K/AKT signaling is associated with carcinogenesis in cancers, especially NKTCL (28,29).
Our study first explored the inhibitory effects of miR-363 on NKTCL and its molecular mechanism. The miR-363 mimic could lead to the inhibition of downstream SIRT6 expression, thereby regulating the PI3K/AKT pathway and inducing the decline of malignant proliferation and apoptosis in most NKTCL cell lines. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5649/rc).
Methods
Clinical samples
To verify the expression of miR-363/SIRT6 in NKTCL, 10 tumor tissue and adjacent normal tissue samples were obtained from NKTCL patients at The Fourth Hospital of Hebei Medical University. All NKTCL patients who only underwent surgery (aged 12 to 77 years) were followed-up until December 2020. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of The Fourth Hospital of Hebei Medical University (No. 2020046) and informed consent was taken from all the patients or the patients’ guardians.
Isolation of normal NK cells from peripheral blood
As previously described (30), the whole-blood samples of healthy blood donors from The Fourth Hospital of Hebei Medical University were prepared to isolate highly purified (90–99%) normal human NK cells using an NK cell separation Kit (Miltenyi Biotec, Germany). The NK cells were then stimulated and cultured in the presence of human recombinant interleukin (IL)-2 (Miltenyi Biotec).
Cell culture and treatments
NKTCL cell lines (SNK-6 and HANK1) were purchased from Shanghai Yaji Biotechnology Co., Ltd. (China). The culture conditions were set according to a previous study (31). When the SNK-6 cells grew to confluence, the cells in each group were transfected with a miR-363 mimic, a mimic negative control (NC), SIRT6-overexpression (OE), or OE-NC using lipo3000 (Life Technologies, USA) for 48 h. The protein and mRNA were then extracted for subsequent expression analysis.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
A qRT-PCR assay was implemented to estimate gene expression. RNA was isolated using a simple total RNA kit (BioTeke, China) and complementary deoxyribonucleic acid (cDNA) was obtained using the Primescript RT Master Mix (TaKaRa, China). The expressions of miR-363 and SIRT6 were quantified by using the SYBR Green Master Mix (TaKaRa). The primer sequences used are listed in Table 1.
Table 1
| Genes | Primer sequence |
|---|---|
| MiR-363 | F: 5'-GCGGCCAATTGCACGGTAT-3' |
| R: 5'-GTGCAGGGTCCGAGGTATTC-3' | |
| U6 | F: 5'-CTCGCTTCGGCAGCACA-3' |
| R: 5'-AACGCTTCACGAATTTGCGT-3' | |
| SIRT6 | F: 5'-TGTGCCAAGTGTAAGACGCAG-3' |
| R: 5'-TTGCCTTAGCCACGGTGCAG-3' | |
| GAPDH | F: 5'-AGACAGCCGCATCTTCTTGT-3' |
| R: 5'-TGATGGCAACAATGTCCACT-3' |
qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot
Total proteins were extracted from cells or tissues using a lysis buffer and then quantified using bicinchoninic acid (BCA) (Pierce, Rockford, IL, USA) to detect the protein expression. Proteins were separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes (Invitrogen, USA) by electroblotting. Subsequently, the membranes were blocked and incubated with the following primary antibodies: SIRT6 (1:1,000, #12486, CST, USA), PI3K (19H8) (1:1,000, #4257, CST, USA), p-PI3K (Tyr458) (1:500, #17366, CST, USA), AKT (C67E7) (1:1,000, #4691, CST, USA), p-AKT (Ser473) (1:500, #4060, CST, USA), and GAPDH (1:2,000, #5174, CST, USA), as well as a secondary antibody immunoglobulin G (IgG) (1:2,000, #14708, CST, USA). Protein bands were visualized using a BeyoECL Star kit (P0018AS, Beyotime, China).
Dual-luciferase reporter assay
A dual-luciferase reporter assay was implemented to affirm the interaction between miR-363 and SIRT6. Briefly, the sequences between miR-363 and SIRT6 (SIRT6-WT) and the respective mutated sequences (SIRT6-MUT) were cloned into pmirGLO vectors (Promega, USA). The SNK-6 cells were transfected with the above-mentioned plasmids, together with miR-363 mimic or NC mimic, and then collected and lysed. Finally, the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) was employed to analyze the luciferase activity (32) post-transfection.
Proliferation analysis
A cell counting kit 8 (CCK-8) experiment was conducted to assess the proliferation ability. Firstly, the SNK-6 cells were inoculated in a 96-well plate. Next, cell viability was examined at specific time points (0, 1, 2, 3, 4, and 5 days) by adding 10 µL of CCK-8 solution to each well. After 4 h of incubation, the microplate reader purchased from Bio-Tek Instruments (Hopkinton, MA, USA) was used to monitor the absorbance at 450 nm.
Apoptosis analysis
To measure cell apoptosis, an AnnexinV-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (C1062M, Beyotime, China) was used to stain the SNK-6 cells. Following removal of the suspended cells, the adherent SNK-6 cells were detached by Trypsin-ethylenediamine tetraacetic acid (EDTA). The SNK-6 cells were suspended in an AnnexinV buffer set and stained with 20 µg/mL propidium iodide (Sigma-Aldrich, USA). The apoptosis of SNK-6 cells was investigated using a flow cytometer (Beckman Coulter, USA), and data were analyzed using the compatible FlowJo software (BD, USA).
Animal experiment
Animal models were established to investigate the effect of miR-363/SIRT6 on NKTCL development in vivo. A total of 28 mice were collected, 24 of which were included in the study due to their healthy status. Four mice were excluded owing to technical or instrumental failures during animal preparation. In the animal experiment, the mice were randomly divided into four groups: mimic NC + OE NC, miR-363 mimic, OE-SIRT6, and miR-363 mimic + OE-SIRT6, with six mice in each group. The 5–6-week-old nude BALB/c male mice (weight 17–20 g) purchased from Charles River Inc. (Beijing) were raised in a specific-pathogen-free (SPF) environment at 26 ℃, with a 12 h day/night cycle at 70% humidity, to eliminate inference.
The subcutaneous transplanted tumor model was established via injection of the transfected SNK-6 (2×107) cells into the right flank skin of the mice (n=6 per group). The tumor volume was recorded every week and calculated according to the following equation: V (mm3) = 0.5 × length × width2, which was constantly monitored during the modeling. Four weeks after inoculation, the mice were sacrificed via carbon dioxide (CO2) inhalation, and thereafter, the tumors were separated and weighed. Animal experiments were performed under a project license (No. 2020047) granted by ethics board of The Fourth Hospital of Hebei Medical University, in compliance with national guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Immunohistochemistry (IHC)
The NKTCL tissue sections (4-µm) were dewaxed and then washed with Tris-buffered saline (TBS) to examine the expression of SIRT6. After soaking and blocking, the sections were incubated with a primary antibody against SIRT6 (1:500, Cruz, sc-517556) overnight at 4 ℃. The next day, the sections were soaked with TBS and Tween-20 (TBST), then incubated with an anti-rabbit secondary antibody for 45 min at 37 ℃, and stained with hematoxylin. After sealing and drying, the positive SIRT6 expression was examined and photographed under a microscope (Olympus, BX63, Japan).
Statistical analysis
All data from triplicated assays were statistically analyzed using GraphPad Prism 8.0 (GraphPad Software, La Jolla, USA), and the results of the three experiments were used for standard deviation (SD) detection. The differences between two or multiple groups were compared using the Student’s t-test or one-way analysis of variation (ANOVA). Data were presented as the mean ± SD as well as the effect size with a 95% confidence interval (CI). P<0.05 was considered statistically significant.
Results
MiR-363 is down-regulated in NKTCL and affects cell proliferation and apoptosis
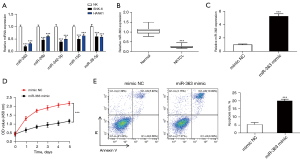
According to Ng et al. (10), among the miRNAs differentially expressed between NKTCL patients and normal controls, the top five down-regulated miRNAs were miR-26b, miR-342-5p, miR-150, miR-363, and miR-28-5p. We verified the expression of these miRNAs in normal NK cells and NK tumor cell lines by qRT-PCR, which showed that miR-363 was most significantly down-regulated in SNK-6 and HANK1 cells (Figure 1A). We found that miR-363 expression was markedly lowly expressed in NKTCL tissues, which is consistent with previous findings (Figure 1B). Further, a mimic NC and miR-363 mimic were transfected into SNK-6 cells, and the transfection efficiency was detected (Figure 1C). CCK-8 and flow cytometry experiments were performed to characterize proliferation and apoptosis of SNK-6 cells in each transfection group. Compared to the control, the miR-363 mimic group showed a significantly reduced SNK-6 cell proliferation level (Figure 1D). As for cell apoptosis, miR-363 overexpression markedly increased the apoptosis capabilities of SNK-6 cells (Figure 1E). These data suggest that miR-363 plays an important role in NKTCL progression by inhibiting the proliferation and promoting the apoptosis of NKTCL cells.
MiR-363 targets SIRT6 to activate the PI3K/AKT pathway
There is evidence that SIRT6 promotes tumorigenesis and drug resistance of DLBCL (25). Herein, the qRT-PCR analysis showed that SIRT6 was notably overexpressed in NKTCL, indicating its potential role (Figure 2A). Also, compared with NK normal cells, we found that SIRT6 was significantly overexpressed in both NKTCL cell lines (Figure 2B). Interestingly, an ENCORI website analysis found that miR-363 and SIRT6 had binding sites (Figure 2C). The dual-luciferase assay conducted in the present study further verified that miR-363 directly regulated the expression of SIRT6 (Figure 2D). Moreover, we observed miR-363 mimic could reduce the mRNA and protein expression of SIRT6 (Figure 2E,2F). Taken together, the above data showed that SIRT6 was up-regulated in NKTCL as a downstream target of miR-363.
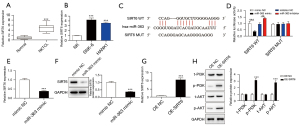
Recent reports have demonstrated that SIRT6 could increase the radiation sensitivity of non-small cell lung cancer (NSCLC) and suppress tumor progression through the PI3K/AKT pathway (33). To better analyze the molecular mechanism of miR-363/SIRT6 participation in NKTCL progression, we further explored the downstream regulatory pathway of SIRT6. So, we overexpressed SIRT6 in SNK-6 cells, and the transfection efficiency was detected (Figure 2G). As indicated in Figure 2H, following the overexpression of SIRT6, the protein expressions of p-PI3K and p-AKT were significantly up-regulated. Taken together, these results suggest that miR-363 could target SIRT6 binding to activate the PI3K/AKT pathway.
MiR-363 affects NKTCL proliferation and apoptosis via the SIRT6/PI3K/AKT pathway in vitro
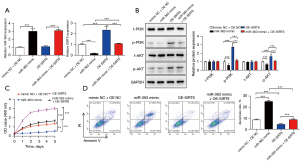
Accordingly, we speculated that the molecular mechanism of miR-363 affecting the progression of NKTCL acted via the SIRT6/PI3K/AKT pathway. To verify this conjecture, we initially transfected SNK-6 cells with a miR-363 mimic or/and OE-SIRT6, and the transfection efficiency was detected by qRT-PCR (Figure 3A). The western blot results showed that p-PI3K and p-AKT were markedly reduced by the miR-363 mimic, which was reversed by OE-SIRT6 transfection (Figure 3B). In addition, CCK-8 and flow cytometry indicated that the miR-363 mimic could substantially restrain cell proliferation and elevate apoptosis, while the effect of OE-SIRT6 on proliferation and apoptosis was opposite to that of the miR-363 mimic. Also, SIRT6 overexpression could notably reverse the influence of the miR-363 mimic on SNK-6 cell proliferation and apoptosis (Figure 3C,3D).
MiR-363 affects NKTCL proliferation and apoptosis via the SIRT6/PI3K/AKT pathway in vivo
To further verify the repressive role of miR-363 on NKTCL in vivo, we inoculated stably transfected SNK-6 cells into nude mice to form transplanted tumors. The tumors in each group were then isolated and photographed (Figure 4A). After 4 weeks, the mean tumor volumes of the miR-363 mimic groups were significantly larger than those of the NC groups. OE-SIRT6 significantly reversed the effect of the miR-363 mimic (Figure 4B). As expected, the tumor weights in each group corresponded to the respective volumes (Figure 4C).
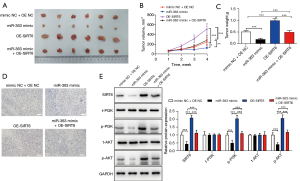
The IHC detection results showed that SIRT6 was substantially decreased in the miR-363 mimic group but increased in the OE-SIRT6 group, and this was reversed in miR-363 + OE-SIRT6 group (Figure 4D). In addition, the protein expression trends of SIRT6, total (t)-PI3K, phosphorylated (p)-PI3K, t-AKT, and p-AKT were consistent with those in vitro (Figure 4E). The in vivo experiments verified that miR-363 affected the progression of NKTCL via the SIRT6/PI3K/AKT pathway.
Discussion
NKTCL is an Epstein-Barr virus (EBV)-related lymphoproliferative disease, accounting for about 2% of T-cell lymphomas. It can be further divided into lymph node (nNKTCL) or extralymph node (eNKTCL), which vary significantly in terms of their clinical, pathophysiological, and genetic characteristics (34). Usually, most eNKTCLs can be seen in the nasal cavity, which is known as nasal-type eNKTCL, but they can be found anywhere. Localized extranodal NKTCL can be cured, but the prognosis of most advanced patients is poor (35,36).
Innate immune cells such as macrophages, neutrophils and natural killer cells and adaptive immune cells can facilitate carcinoma process in tumor microenvironment, lymphoma contained (37,38). MiRNAs have been demonstrated to regulate microenvironment or function in microenvironment during tumor progression (39,40). Moreover, numerous studies have confirmed that miRNAs possess the ability of multi-gene regulation to directly or indirectly regulate various signaling pathways and thereby affect the occurrence and metastasis of malignant tumors (41). For instance, miR-155 has been illustrated to target BRG1 to activate STAT3/VEGFC signaling and mediate lymphangiogenesis in NKTCL (42). And miR-188-5p refrains NKTCL progression by inhibited regulation of XRCC5 (43). Thus, we attempted to dig out functional miRNAs in NKTCL. After browsing website, we uncovered the top five down-regulated miRNAs, miR-26b, miR-342-5p, miR-150, miR-363, and miR-28-5p contained, in NKTCL patients (10). After verification, it was observed that miR-363 exhibited the most significant down-regulation in SNK-6 and HANK1 cells, suggesting the involvement of miR-363 in NKTCL. According to previous studies, miR-363 is a typical and effective tumor-suppressor gene in cancer (44,45), which is consistent with our research. Furthermore, miR-363 has been indicated to regulate SQLE that is significantly related with tumor immune cell infiltration and checkpoints in pancreatic adenocarcinoma (46). However, miR-363 has been rarely reported in NKTCL. Therefore, the present study aimed to reveal the significance and mechanism of miR-363 in NKTCL for the first time.
Herein, we found that miR-363 was down-regulated in NKTCL tissues and cell lines, which affected the proliferation and apoptosis of SNK-6 cells. It is well known that miRNAs with tumor-suppressive functions are abnormally lowly expressed in tumors and can regulate the growth and metastasis of cancer cells by targeting and up-regulating the expression of downstream tumor-related genes (47). Given this, we further studied the downstream regulatory mechanism of miR-363. Among its many target genes, SIRT6 attracted our attention.
Abnormal SIRT6 expression is associated with a variety of malignant tumors, such as gastric cancer (48), ovarian cancer (49), medulloblastoma (50), etc. There is also evidence that SIRT6 can be regarded as an effective tumor suppressor owing to its effective regulation of aerobic glycolysis of cancer cells, which suppresses tumor metabolism (17). Considering the primary role of SIRT6 in intracellular homeostasis, it has broad prospects for development into small molecule inhibitors or activators and has emerging therapeutic potential for cancer diagnosis and treatment (51). Yet, the role and mechanism of SIRT6 in NKTCL have not been reported.
In our study, we firstly elucidated the significant up-regulation of SIRT6 in NKTCL clinical tissues, and qRT-PCR experiments demonstrated that SIRT6 was significantly overexpressed in NKTCL cell lines. Moreover, the rescue experiments showed that miR-363 could target SIRT6 binding to activate the PI3K/AKT pathway. It is common for constitutively activating signaling pathways to regulate the development of malignant tumors (52). Furthermore, the amplification of PI3K/AKT signaling has been shown to mediate the occurrence of a variety of cancers (53).
To verify whether miR-363 affects the progression of NKTCL via the SIRT6/PI3K/AKT pathway axis, cellular and mouse tumor formation experiments were carried out. The results revealed that the differential expression of miR-363 or SIRT6 significantly affected the activation of the PI3K/AKT pathway, and thus, changed the proliferation and apoptosis levels of NKTCL cell lines. Moreover, the miR-363/SIRT6/PI3K/AKT axis mechanism affecting mouse NKTCL progression was also confirmed, and the data trends were consistent with SNK-6 cells.
However, there are several limitations in the current research that should be noted. Firstly, the potential upstream molecule of the miR-363/SIRT6 axis in NKTCL was not investigated. Also, other biological functions, such as stemness, migration, invasion, etc., were not analyzed. These considerations will be included in our future studies.
Conclusions
Expression of miR-363 in NKTCL is significantly lower than that in normal tissues or cells. MiR-363 could repress proliferation but accelerate apoptosis via targeting SIRT6 to activate PI3K/AKT pathway in NKTCL, which is expected to provide a new target and guidance for the treatment and diagnosis of NKTCL, as well as offer an additional molecular option for future drug development of NKTCL.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5649/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5649/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5649/coif). The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal protocols in this study were performed under a project license (No. 2020047) granted by ethics board of The Fourth Hospital of Hebei Medical University, in compliance with national guidelines for the care and use of animals. Experiments involving human participants were approved by the ethics board of The Fourth Hospital of Hebei Medical University (No. 2020046), and was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients or the patients’ guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cai Q, Luo X, Zhang G, et al. New prognostic model for extranodal natural killer/T cell lymphoma, nasal type. Ann Hematol 2014;93:1541-9. [Crossref] [PubMed]
- Anderson JR, Armitage JO, Weisenburger DD. Epidemiology of the non-Hodgkin's lymphomas: distributions of the major subtypes differ by geographic locations. Non-Hodgkin's Lymphoma Classification Project. Ann Oncol 1998;9:717-20. [Crossref] [PubMed]
- Nakamura S, Koshikawa T, Koike K, et al. Phenotypic analysis of peripheral T cell lymphoma among the Japanese. Acta Pathol Jpn 1993;43:396-412. [Crossref] [PubMed]
- Bi XW, Li YX, Fang H, et al. High-dose and extended-field intensity modulated radiation therapy for early-stage NK/T-cell lymphoma of Waldeyer's ring: dosimetric analysis and clinical outcome. Int J Radiat Oncol Biol Phys 2013;87:1086-93. [Crossref] [PubMed]
- Wang L, Wang ZH, Chen XQ, et al. First-line combination of gemcitabine, oxaliplatin, and L-asparaginase (GELOX) followed by involved-field radiation therapy for patients with stage IE/IIE extranodal natural killer/T-cell lymphoma. Cancer 2013;119:348-55. [Crossref] [PubMed]
- Jeong SH. Extranodal NK/T cell lymphoma. Blood Res 2020;55:S63-71. [Crossref] [PubMed]
- Plank M, Maltby S, Mattes J, et al. Targeting translational control as a novel way to treat inflammatory disease: the emerging role of microRNAs. Clin Exp Allergy 2013;43:981-99. [Crossref] [PubMed]
- Fernández-Hernando C, Ramírez CM, Goedeke L, et al. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol 2013;33:178-85. [Crossref] [PubMed]
- Saki N, Abroun S, Soleimani M, et al. Involvement of MicroRNA in T-Cell Differentiation and Malignancy. Int J Hematol Oncol Stem Cell Res 2015;9:33-49. [PubMed]
- Ng SB, Yan J, Huang G, et al. Dysregulated microRNAs affect pathways and targets of biologic relevance in nasal-type natural killer/T-cell lymphoma. Blood 2011;118:4919-29. [Crossref] [PubMed]
- Yang C, Han S. The circular RNA circ0005654 interacts with specificity protein 1 via microRNA-363 sequestration to promote gastric cancer progression. Bioengineered 2021;12:6305-17. [Crossref] [PubMed]
- Ren L, Zhou H, Lei L, et al. Long non-coding RNA FOXD3 antisense RNA 1 augments anti-estrogen resistance in breast cancer cells through the microRNA-363/ trefoil factor 1/ phosphatidylinositol 3-kinase/protein kinase B axis. Bioengineered 2021;12:5266-78. [Crossref] [PubMed]
- Gowda PS, Wildman BJ, Trotter TN, et al. Runx2 Suppression by miR-342 and miR-363 Inhibits Multiple Myeloma Progression. Mol Cancer Res 2018;16:1138-48. [Crossref] [PubMed]
- Dong J, Geng J, Tan W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed Pharmacother 2018;105:922-31. [Crossref] [PubMed]
- Mohamed Z, Hassan MK, Okasha S, et al. miR-363 confers taxane resistance in ovarian cancer by targeting the Hippo pathway member, LATS2. Oncotarget 2018;9:30053-65. [Crossref] [PubMed]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 2009;460:587-91. [Crossref] [PubMed]
- Sebastián C, Zwaans BM, Silberman DM, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012;151:1185-99. [Crossref] [PubMed]
- Marquardt JU, Fischer K, Baus K, et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology 2013;58:1054-64. [Crossref] [PubMed]
- Bai L, Lin G, Sun L, et al. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget 2016;7:40377-86. [Crossref] [PubMed]
- Kugel S, Sebastián C, Fitamant J, et al. SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell 2016;165:1401-15. [Crossref] [PubMed]
- Ran LK, Chen Y, Zhang ZZ, et al. SIRT6 Overexpression Potentiates Apoptosis Evasion in Hepatocellular Carcinoma via BCL2-Associated X Protein-Dependent Apoptotic Pathway. Clin Cancer Res 2016;22:3372-82. [Crossref] [PubMed]
- Liu Y, Xie QR, Wang B, et al. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell 2013;4:702-10. [Crossref] [PubMed]
- Ruan L, Chen J, Ruan L, et al. miR-34a inhibits tumorigenesis of NSCLC via targeting SIRT6. Int J Clin Exp Pathol 2018;11:1135-45. [PubMed]
- Ruan ZF, Xie M, Gui SJ, et al. MiR-370 accelerated cerebral ischemia reperfusion injury via targeting SIRT6 and regulating Nrf2/ARE signal pathway. Kaohsiung J Med Sci 2020;36:741-9. [Crossref] [PubMed]
- Yang J, Li Y, Zhang Y, et al. Sirt6 promotes tumorigenesis and drug resistance of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J Exp Clin Cancer Res 2020;39:142. [Crossref] [PubMed]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002;2:489-501. [Crossref] [PubMed]
- Uddin S, Hussain AR, Al-Hussein KA, et al. Inhibition of phosphatidylinositol 3'-kinase/AKT signaling promotes apoptosis of primary effusion lymphoma cells. Clin Cancer Res 2005;11:3102-8. [Crossref] [PubMed]
- Noorolyai S, Shajari N, Baghbani E, et al. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019;698:120-8. [Crossref] [PubMed]
- Wendel HG, De Stanchina E, Fridman JS, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 2004;428:332-7. [Crossref] [PubMed]
- Ng SB, Selvarajan V, Huang G, et al. Activated oncogenic pathways and therapeutic targets in extranodal nasal-type NK/T cell lymphoma revealed by gene expression profiling. J Pathol 2011;223:496-510. [Crossref] [PubMed]
- Lin R, Li X, Wu S, et al. Suppression of latent transforming growth factor-β (TGF-β)-binding protein 1 (LTBP1) inhibits natural killer/ T cell lymphoma progression by inactivating the TGF-β/Smad and p38MAPK pathways. Exp Cell Res 2021;407:112790. [Crossref] [PubMed]
- Shang J, Sun S, Zhang L, et al. miR-211 alleviates ischaemia/reperfusion-induced kidney injury by targeting TGFβR2/TGF-β/SMAD3 pathway. Bioengineered 2020;11:547-57. [Crossref] [PubMed]
- Xiong L, Tan B, Lei X, et al. SIRT6 through PI3K/Akt/mTOR signaling pathway to enhance radiosensitivity of non-Small cell lung cancer and inhibit tumor progression. IUBMB Life 2021;73:1092-102. [Crossref] [PubMed]
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. Blood 2016;128:462-3. [Crossref] [PubMed]
- Kim SJ, Yoon SE, Kim WS. Treatment of localized extranodal NK/T cell lymphoma, nasal type: a systematic review. J Hematol Oncol 2018;11:140. [Crossref] [PubMed]
- Yang Y, Zhu Y, Cao JZ, et al. Risk-adapted therapy for early-stage extranodal nasal-type NK/T-cell lymphoma: analysis from a multicenter study. Blood 2015;126:1424-32; quiz 1517. [Crossref] [PubMed]
- Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res 2019;79:4557-66. [Crossref] [PubMed]
- Xu ML, Fedoriw Y. Lymphoma Microenvironment and Immunotherapy. Surg Pathol Clin 2016;9:93-100. [Crossref] [PubMed]
- Zhang Y, Huo W, Sun L, et al. Targeting miR-148b-5p Inhibits Immunity Microenvironment and Gastric Cancer Progression. Front Immunol 2021;12:590447. [Crossref] [PubMed]
- Neviani P, Wise PM, Murtadha M, et al. Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res 2019;79:1151-64. [Crossref] [PubMed]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014;9:287-314. [Crossref] [PubMed]
- Chang Y, Cui M, Fu X, et al. MiRNA-155 regulates lymphangiogenesis in natural killer/T-cell lymphoma by targeting BRG1. Cancer Biol Ther 2019;20:31-41. [Crossref] [PubMed]
- Huang Q, Ding S, Zhang H. Regulatory effects of miR-188-5p/XRCC5 on the progression of natural killer/T-cell lymphoma. J BUON 2021;26:2033-9. [PubMed]
- Li G. Expression of RUNX3 gene and miR-363 in colorectal cancer and the relationship with clinicopathological features. Oncol Lett 2019;18:2278-85. [Crossref] [PubMed]
- Lou W, Ding B, Zhong G, et al. Dysregulation of pseudogene/lncRNA-hsa-miR-363-3p-SPOCK2 pathway fuels stage progression of ovarian cancer. Aging (Albany NY) 2019;11:11416-39. [Crossref] [PubMed]
- You W, Ke J, Chen Y, et al. SQLE, A Key Enzyme in Cholesterol Metabolism, Correlates With Tumor Immune Infiltration and Immunotherapy Outcome of Pancreatic Adenocarcinoma. Front Immunol 2022;13:864244. [Crossref] [PubMed]
- Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int 2015;15:38. [Crossref] [PubMed]
- Cai S, Fu S, Zhang W, et al. SIRT6 silencing overcomes resistance to sorafenib by promoting ferroptosis in gastric cancer. Biochem Biophys Res Commun 2021;577:158-64. [Crossref] [PubMed]
- Wang H, Li J, Huang R, et al. SIRT4 and SIRT6 Serve as Novel Prognostic Biomarkers With Competitive Functions in Serous Ovarian Cancer. Front Genet 2021;12:666630. [Crossref] [PubMed]
- Zhu C, Li K, Jiang M, et al. RBM5-AS1 promotes radioresistance in medulloblastoma through stabilization of SIRT6 protein. Acta Neuropathol Commun 2021;9:123. [Crossref] [PubMed]
- Fiorentino F, Mai A, Rotili D. Emerging Therapeutic Potential of SIRT6 Modulators. J Med Chem 2021;64:9732-58. [Crossref] [PubMed]
- Benekli M, Baer MR, Baumann H, et al. Signal transducer and activator of transcription proteins in leukemias. Blood 2003;101:2940-54. [Crossref] [PubMed]
- Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin Cancer Biol 2019;59:147-60. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


