
A deep learning model using convolutional neural networks for caries detection and recognition with endoscopes
Highlight box
Key findings
• A deep learning model with classification and segmentation parts to detect caries lesions in non-clinical settings like schools or communities or the household.
What is known and what is new?
• We have built a deep learning model, by which we can not only determine whether there is caries, but also obtain the areas of caries;
• We have added a segmentation model to directly show the areas of the lesion with endoscopes, a more conveniently used tool.
What is the implication, and what should change now?
• The deep learning model can be used in non-clinical situations, but we still need more pictures to improve the accuracy and sensitivity of the model, what’s more, we could build a platform based on our deep learning algorithm to help people diagnose the caries by themselves.
Introduction
Caries are one of the world’s most common chronic diseases (1). Some studies have demonstrated that the number of untreated caries worldwide is about 2.3 billion; in some economically developed countries, the number of cases and their burden are lower than those in low-income countries (2). Currently, caries are diagnosed by inspection and probing, sometimes using dental radiography (3). However, in some low- and lower-middle-income countries, a lack of dentists and unequal distribution of medical resources (4)—most of which are concentrated in big cities—can make it difficult for ordinary people to seek regular medical care. Substandard awareness of oral hygiene is another frequent problem within this population, sometimes they cannot even realize that they have oral problems until they feel obvious pain, which, is too late. As emergency room visits declined during the coronavirus disease of 2019 (COVID-19) epidemic, the proportion of patients exhibiting inflammation increased (5). Situations like these place additional pressure on dentists and patients because of insufficient medical resources. Taken together, these factors may increase the likelihood of missing the treatment window.
In recent years, deep learning—and especially convolutional neural networks (CNNs)—have been widely adopted for medical image analysis (6). As a data-driven algorithm, deep learning model has a strong ability to fit distribution through backpropagation algorithm, it can adaptively extract task-related features with multiple layers that can learn representations of data and has a strong generalization ability. These networks convert images into data to facilitate clinical diagnosis and decision making (7). For example, several oncology studies have found that deep learning could extract images from computed tomography (CT) and magnetic resonance imaging (MRI) (8-10). In dentistry, CNNs have be applied to various fields (11) such as periodontal diseases (12) and orthodontics (13). Although some studies have focused on caries, the deep learning models that they used have usually been based on near-infrared transillumination images (14) and radiographic images (15), and seldom on camera-based images (16). These methods have high diagnostic accuracy and support the potential of deep learning; however, they are mostly used in clinical settings and cannot, therefore, relieve the pressure on healthcare providers in regions with insufficient medical resources.
To address this problem, we produced a kind of endoscope which is cheap, convenient, and can be connected to mobile phones. Its camera is small enough to be placed in the mouth and take photos of your teeth. Further, we chose to use a deep learning algorithm that could directly detect and recognize the area of caries by using 2 parts. The former is a classification part to detect whether there are caries and the latter is a segmentation model to label and directly show the areas of the lesion. This might help patients to better understand their disease and maintain good oral hygiene to prevent caries from worsening, even in low- and lower-middle-income countries. Compared to the mobile phones, there is no need for one endoscope per person, it can be used in a family or even a school or a village. In addition, deep learning model is a data-driven algorithm with strong ability of fitting distribution, which means it can get the same accuracy as the model image-taken by cameras. Therefore, this study aimed to establish and evaluate a deep learning model for detecting and recognizing areas with caries using endoscopy. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5816/rc).
Methods
The study mainly includes the establishment of the database, preprocessing and image augmentation, the architecture of the deep CNN algorithm and the evaluation of the model.
Database
The dataset consisted of 1,059 caries images from permanent molars and premolars and 194 images of non-caries in patients. All the images were obtained by endoscopes through the Department of Stomatology, Chinese PLA General Hospital. All patients who participated in this study signed an informed consent form, and this study was approved by the Ethics Committee of Chinese PLA General Hospital (No. S2019-016-02). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). As for the data collector, we decided not to train the users to better simulate the patient’s own photo-taking situation. The endoscopes could be connected to mobile phones with Wi-Fi for image viewing using an app called JVE (Huakangzhongjian Co., Ltd, China). All caries images in this study were acquired between 2019 and 2022. We collected the data by people who did not be trained, such as patients in outpatient or intern students, to make sure that when users took photos by themselves under non-medical situations. In this way, we could avoid the bias caused by the difference of image quality between different situations.
The golden criteria for caries were as follows: (I) the areas which appeared as black or white spots. (II) Cavitated lesions that felt rough or conveyed pain or discomfort upon probing. (III) Temporary fillings or amalgam fillings that had been broken (17).
All images were labeled using Labelme (Labelme.4.5.6), including 2 parts: (I) the whole area: only the teeth in the pictures were labeled as the background. (II) The lesions: the areas with black or white spot lesions and those areas that were probed roughly or with sticking. Two experienced dentists worked independently, with a third expert intervening in cases of disagreement. Since the number of images was insufficient for splitting into the fixed training and test datasets, we finally chose the 5-fold cross-validation protocol. Each fold was used once as the validation and the remaining folds formed the training set. We shuffled all images and divided them into 5 parts using Scikit-learn, an open-source machine learning library.
In task 1, we used the ResNet101 neural network to identify positive and negative samples. ResNet101 consists of several ResBlocks, which use a 3×3 convolution layer, a 1×1 convolution layer, and a 3×3 convolution layer. The shortcut pathway will add inputs to outputs directly. In task 2, we designed a segmentation model. The segmentation model is an encoder-decoder architecture, where the encoder part takes ResNet101 with SE block as the backbone network. The atrous spatial pyramid pooling module (ASPP) is used to extract multi-scale context information. The decoder part outputs final segmentation results by combining and upsampling multi-level features.
Preprocessing and image augmentation
First, we collected 194 normal camera images as negative lesion classification data and chose 194 caries lesion images as positive classification part data from the total 1,059 lesion images randomly. In this way we can avoid the imbalance caused by the large difference of images numbers between the two sets of data We also employed the same 5-fold validation protocol and used enhancement to avoid overfitting. In the classification task, we first used RandomResizedCrop [scale = (0.5, 1.0); ratio = (0.75, 1.33)], but we resized the crop to 224×224 pixels. Before normalizing the image, we randomly changed the brightness and contrast of the input image using RandomBrightnessContrast (P=0.5).
To evaluate the effectiveness of our segmentation method, we collected 1,059 camera images which were converted into PNG files. To avoid overfitting, the training dataset was augmented by RandomResizedCrop [scale = (0.5, 1.0), ratio = (0.75, 1.33)]. Specifically, a crop of the random size of the original size and a random aspect ratio of the original aspect ratio was made. Then, this crop was resized to 512×512 pixels. Finally, training images were normalized into (−1, 1). The whole process is shown in Figure 1.
Architecture of the deep CNN algorithm
ResNet101—a CNN equipped with residual architecture—was used to identify negative examples for the lesion classification task. This was different from previous plain CNNs, which only stacked convolution layers sequentially. The residual CNN demonstrated shortcuts between stacked convolution layers. Because of these additional pathways, the gradients could be easily transferred to any layer. This allowed us to train a deeper CNN. ResNet101 was used as a classification network and as a backbone network for subsequent segmentation models in this paper. Specifically, ResNet101 identified approximately 101 convolution layers. After the global average pool, a one-dimensional feature was created for the final prediction. We choose the binary cross entropy loss as classification loss; the training settings were the same as those for the following segmentation task.
An end-to-end semantic segmentation model—DeepLabv3+ (18)—was used for segmenting the dental lesions. It was an encoder-decoder structure network where the encoder stage extracted multi-scale object contextual information using a backbone network and atrous convolution. During the decoder stage, features from multiple levels were fused and the segmentation results were refined along object boundaries. Specifically, ResNet101 (19) with SE block (20) was used as the backbone network, pre-trained by the ImageNet (21) dataset. Then, the ASPP module (22)—with different atrous rates—was used to capture information across multiple scales while keeping the encoded feature map its original size. At the decoder stage, encoded features were upsampled and concatenated with corresponding low-level features from the backbone network. Several convolutions and another upsampling procedures were used to refine the features and form final predictions. Cross-entropy loss, Dice loss (23), and intersection over union (IoU) loss (24) were collectively used to train the segmentation model. We adopted stochastic gradient descent (SGD) optimization with momentum =0.9, weight decay =0.0001, initial learning rate =0.01, and multiplied it by 0.1 at 180 epoch and 250 epoch. We trained a network consisting of 300 epochs with 8 Nvidia Titan Xp GPUs (Graphics Processing Units); each GPU had a batch size of 8.
Statistical analysis
First, we used accuracy, recall and receiver operating characteristic (ROC) curve, and area under the ROC curve (AUC) to assess the classification model. P statistical significance was indicated when P<0.01, and 90% confidence intervals (CIs) were calculated. After this, we chose accuracy, recall, specificity, Dice, IoU, positive predictive value (PPV), and negative predictive value (NPV) to evaluate the segmentation model by comparing manual-labeled areas with predicted areas.
True positive (TP) means the part which is judged positive and actually positive, false positive (FP) means the part which is judged positive but actually negative, true negative (TN) means the part which is judged negative and actually negative, false negative (FN) means the part which is judged negative but actually positive.
Accuracy was the ratio of the actual positive parts to all positive parts.
Recall was the ratio of the TP samples in actual positive samples.
Specificity was the ratio of TN samples in actual negative samples.
PPV was the ratio of TP samples in predicted positive samples.
NPV was the ratio of TN samples in predicted negative samples.
The Dice similarity coefficient (DSC) gauges the similarity of 2 sets, which means that the 2 sets’ (A and B), DSC is defined as . In this paper, we supposed that set A is a true lesion area on a tooth, and set B is the segmentation area that the model predicts. The numerator means TP, and A+B can be represented as the union of , where the latter 2 items represent FN and FP. Therefore, DSC can also be represented as . The DSC is popular in semantic segmentation tasks for its simplicity; we also used it as a component of our evaluation system. As for the statistical software, we used pytorchlightning toolbox based on Pytorch framework to do the work.
Results
As shown in Table 1, the mean accuracy was 0.9637 [range, 0.9489–0.9854, standard deviation (SD) =0.0152], the recall was 0.9796 (range, 0.9583–0.9903, SD =0.0132). TN =33, 40, 35, 31, 40; TP =102, 92, 96, 99, 95; FP =2, 2, 4, 6, 1; FN =1, 4, 3, 1, 1. The ROC curves for the 5 folds of the classification model are shown in the Figure 2, The mean AUC was 0.9897 (range, 0.9821–0.9956, SD =0.00564). Statistical significance was indicated when P<0.01, and 90% CIs were calculated.
Table 1
| Variables | Fold0 | Fold1 | Fold2 | Fold3 | Fold4 | Mean | SD |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.9783 | 0.9565 | 0.9493 | 0.9489 | 0.9854 | 0.9637 | 0.0152 |
| Recall | 0.9903 | 0.9583 | 0.9697 | 0.9900 | 0.9896 | 0.9796 | 0.0132 |
| TN | 33 | 40 | 35 | 31 | 40 | – | – |
| TP | 102 | 92 | 96 | 99 | 95 | – | – |
| FP | 2 | 2 | 4 | 6 | 1 | – | – |
| FN | 1 | 4 | 3 | 1 | 1 | – | – |
| AUC | 0.9783 | 0.9565 | 0.9493 | 0.9489 | 0.9854 | 0.9897 | 0.0152 |
AUC, area under the curve; TN, true negative; TP, true positive; FP, false positive; FN, false negative; SD, standard deviation.
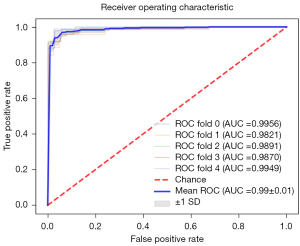
In Table 2, the mean accuracy was 0.9843 (range, 0.9820–0.9871, SD =0.0016), the recall was 0.6996 (range, 0.6810–0.7194, SD =0.0124), the specificity was 0.9943 (range, 0.9937–0.9954, SD =0.0006), the Dice coefficient was 0.7099 (range, 0.6948–0.7343, SD =0.0143), the IoU is 0.5779 (range, 0.5646–0.6006, SD =0.0138), the PPV was 0.7804 (range, 0.7634–0.8023, SD =0.0128), the NPV was 0.9887 (range, 0.9866–0.9909, SD =0.0014).
Table 2
| Variables | Fold0 | Fold1 | Fold2 | Fold3 | Fold4 | Mean | SD |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.9839 | 0.9838 | 0.9820 | 0.9871 | 0.9845 | 0.9843 | 0.0016 |
| Recall | 0.7002 | 0.6810 | 0.6945 | 0.7028 | 0.7194 | 0.6996 | 0.0124 |
| Specificity | 0.9940 | 0.9937 | 0.9946 | 0.9954 | 0.9940 | 0.9943 | 0.0006 |
| Dice | 0.7038 | 0.6948 | 0.6992 | 0.7172 | 0.7343 | 0.7099 | 0.0143 |
| IoU | 0.5699 | 0.5646 | 0.5674 | 0.5870 | 0.6006 | 0.5779 | 0.0138 |
| PPV | 0.7739 | 0.7797 | 0.7634 | 0.7829 | 0.8023 | 0.7804 | 0.0128 |
| NPV | 0.9883 | 0.9885 | 0.9866 | 0.9909 | 0.9892 | 0.9887 | 0.0014 |
IoU, intersection over union; PPV, positive predictive value; NPV, negative predictive value; SD, standard deviation.
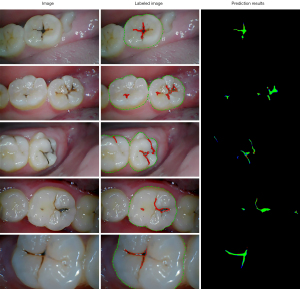
Figure 3 includes predicted results and original images, labeled to show the ability of our deep learning model to recognize the areas containing caries lesions. We could indicate that the segmentation model can display the lesions in a more audio-visual way than showing the original pictures simply. It is worth mentioning that we determined the caries’ teeth as background area, so the black parts only in teeth with lesions mean the negative areas, rather than these of the whole picture.
Discussion
Early discovery and diagnosis of caries are important (25) for preventing irreversible damage and severe pain. However, in some economically undeveloped regions, diagnosis and treatment of caries may be limited by the lack of medical resources (26). To solve this problem, researchers have used deep learning models to recognize caries. However, the past studies of deep learning models for dental caries have used periapical radiographic images or near-infrared transillumination images (27,28). These methods are both accurate and sensitive but seldom can be used in non-clinical settings like schools or communities or the household. Considering this, we sought to build a deep learning model based on endoscopic images with high generality and popularity, meaning that it cannot be used in medical situations, is relatively inexpensive, and easy to operate. The model consisted of 2 parts: classification part and segmentation part. The former can diagnose the presence of caries and the latter can detect and manifest its area. We aimed to find a more direct vision to understand the patients their lesion by adding a segmentation model.
As shown in Table 1, the mean accuracy of classification model was 0.9637, the mean recall was 0.9796, the mean AUC was 0.9897, showing a considerably good performance in detecting the caries. These results met the standard that detected whether there was caries lesion. However, we wanted to find a more visible way to enable patients to understand their situation. Therefore, we decided to add a segmentation model.
As shown in Table 2, the average segmentation accuracy of segmentation model was 0.9843, which was considered too high. An overly high TN value may conceal significant disagreement between the segmented images and predictions. We had a similar problem with specificity (the average was 0.9943). This is because the caries lesion areas are only a small part of the picture, although we had already chosen the teeth areas as the background. To solve this problem, we used random crop to augment the dataset. We also revealed that the Dice score and IoU for identifying dental caries are above 0.7099 and 0.5779, respectively. Besides this, the recall and PPV for identifying dental caries were integrated to evaluate the model’s effectiveness. We summarized the possible reasons for the results as follows: firstly, lack of enough training images would fail to effectively train the deep learning model and potentially influence the model’s accuracy. Secondly, labeling also produces mistakes because caries may be small on the top yet big on the bottom and the edge of it may also be blurred. These layers can blend together, render the edge non-specific, and thus increase the difficulty of labeling. Thirdly, the area of the caries would only include a little part of the whole background; even a minor difference could markedly affect the patient’s outcome, which is a common problem in segmentation deep learning model, especially where the lesion areas make up a small prat of the whole picture. Therefore, the model may not have sufficient training for recognizing caries.
Unlike traditional machine learning or artificial neural networks, CNNs stack multiple convolution layers, pooling layers, and fully connected layers as the backbone network to extract abstract and informative features. Then, task-specific lightweight networks use those powerful features to solve specific visual recognition tasks such as image classification, object detection, or semantic segmentation. ResNet, a popular image classification method, uses residual architecture to successfully alleviate the degradation problem inherent to deeper networks. ResNet quickly caught the attention of experts and researchers and was eventually applied to various fields after being shown to be effective. ResNet has a simple structure consisting of convolutional layers, some fully connected layers, and a global average pooling layer. The stacked convolutional layers continuously abstract the valid information in the data. The global average pooling layer obtains global information. Finally, the image features are mapped to the category space by the fully connected layer for image classification. In this paper, we identified positive and negative cases based on a 101-layer ResNet network.
Based on residual architecture, lots of deeper or denser connection deep learning algorithms, such as SENet and DenseNet (29), are continually being proposed and used as the backbone networks for improving the performance of other vision tasks. As a classical segmentation model, DeepLabv3+ can be used to solve the lesion segmentation task. Benefitting from the ASPP module and multi-level feature upsample and fusion, the DeepLabv3+ network can extract multi-scale features without reducing the resolution of the output feature maps. Therefore, DeepLabv3+—which takes ResNet with SE blocks as the powerful backbone network—can achieve satisfying performance. In addition, continually progressing deep learning algorithms and a huge amount of accessible open-source code can increasingly improve future task performance.
A key feature of our study was the selection of endoscopes as our imaging tool. Endoscopes are low-cost, small, and easy-to-use tools. A portable endoscope could be used in non-clinical settings, but with sufficient definition to identify and recognize caries. Such technology would be easier to deploy, potentially reducing pressure in areas with limited medical resources. Endoscopes can not only obtain photos which can hardly be seen by the eyes, such as maxillary molars, but also capture 1 face of a tooth at a time. This allowed observation of all the blind angles of teeth, allowing inspection of the entire oral cavity. The image resolution provided by endoscopes can meet the standard of the deep learning model to recognize areas containing caries.
The deep learning model can potentially identify the caries area accurately for two fundamental reasons. One is from the point of view of data: because of the portable endoscopes, the generalization performance of traditional digital image processing methods is poor due to the light, shooting angle, impurities and imaging quality, etc. Second, from the perspective of model, deep learning model is a data-driven algorithm with strong ability of fitting distribution. Through backpropagation algorithm, task-related features can be extracted adaptively, with strong generalization ability. Additionally, we added a segmentation part to improve the deep learning model, in some economically undeveloped areas, those who lack awareness of oral health care still cannot distinguish the difference among caries, pigment, and dental calculus, even if you show them the original endoscopes images. Figure 4 shows the primary output image shown to the users of this segmentation model; in the next stage we would like to overlap the labeled area and original pictures to further educate the users. By using segmentation model, patients can be shown labeled pictures of their caries, potentially encouraging them to seek prompt medical attention. In addition, people using this deep learning model can build a dental health record by capturing and uploading pictures regularly. In this way, we can compare the pictures with their last times’, and monitor any change of caries lesions.
Our study also had several limitations. First, and most notably, the limited number of trained images included in our deep learning model may not have conveyed sufficient training. Secondly, our experiment included several interference factors, including the environment and teeth factors. The former includes different light degrees, specular reflection, saliva, the fog on the mirror, and imaging instability. The latter includes pigment, dental calculus, white spot lesions, food impaction, past dental therapy (silver amalgam or orthodontic brackets). Otherwise, we only focused on fissure caries in premolars and molars because we lacked data from other kinds of caries, such as proximal dental caries. Additionally, some caries may be small on the top but larger on the bottom. These layers blend together so that the edges become difficult to identify. Therefore, our model may not recognize deep lesions under the tooth’s surface, meaning we cannot effectively identify occult caries. We can only obtain images of the tooth’s surface. For these reasons, the model now cannot replace the human doctors to accurately diagnosed the caries, but it can help people achieve early detection of caries lesion.
Future efforts should build upon our work. Importantly, we will continue to collect more images—and images of teeth other than premolars and molars—to extend our database. This will improve the accuracy of our predictive model. Besides this, we plan to create an auto-labeled algorithm to label the input pictures. After being labeled by the algorithm, researchers will check and modify them. In this way, we can have a more efficient model as well. Meanwhile, we will continue to study the influence of interference factors and strategies to minimize their impact, for instance, an algorithm that can segregate the interference areas and the lesion areas. In the future, we can build additional deep learning models to diagnose other oral diseases, such as periodontitis, white spot lesions, mucosal disease, or even oral cancers. Given uneven network coverage patterns, we will further optimize our model to reduce its resource consumption (storage, computational power, etc.). This would allow it to run on low-cost endpoints such as endoscopes or mobile terminals, serving additional people. In addition, since endoscopes are handheld devices and are widely used by novices, our model was affected by inclusion of blurred images and the relative scarcity of lesion areas in our collected data; these could affect the final detection results. In the future, we can also create an artificial intelligence platform based on our deep learning algorithm. Patients may be able to upload images captured by endoscopes and receive a diagnosis from our deep learning model. When patients receive labeled images back from the platform, they can review the image again and make any additional comments. If possible, images will be conveyed to the background system and reevaluated by our team of doctors. The new images will be sent to the patients as well as to the deep learning model for further training and corrections. This should improve the platform’s accuracy, potentially increasing its uptake with patients. In the future, computing time may be reduced by using lightweight deep learning models to achieve real-time detection through video and avoid low lesion detection rates from single-frame images taken by novice endoscopists. This method may help relieve pressure on dentists, particularly in times of medical resource rationing or in areas with limited access to healthcare services. Patients then may be able to be diagnosed using images obtained at home using endoscopes and then uploaded to our platform.
Conclusions
We used a deep learning model to develop an easy-to-use and relatively precise way for people to monitor and detect caries. This model, though, is not a complete replacement for doctors for its accuracy, our method may facilitate early discovery and diagnosis of caries lesions and provide an easy and low-cost means of managing oral health, particularly in underserved areas.
Acknowledgments
Funding: This research was funded by the National Key R&D Program of China (No. 2020YFC2008900).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5816/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5816/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5816/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients who participated in this study signed an informed consent form, and this study was approved by the Ethics Committee of Chinese PLA General Hospital (No. S2019-016-02). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kassebaum NJ, Bernabé E, Dahiya M, et al. Global burden of untreated caries: a systematic review and metaregression. J Dent Res 2015;94:650-8. [Crossref] [PubMed]
- Lagerweij MD, van Loveren C. Declining Caries Trends: Are We Satisfied? Curr Oral Health Rep 2015;2:212-7. [Crossref] [PubMed]
- Fee PA, Macey R, Walsh T, et al. Tests to detect and inform the diagnosis of root caries. Cochrane Database Syst Rev 2020;12:CD013806. [Crossref] [PubMed]
- GBD 2017 Oral Disorders Collaborators, Bernabe E, Marcenes W, et al. Global, Regional, and National Levels and Trends in Burden of Oral Conditions from 1990 to 2017: A Systematic Analysis for the Global Burden of Disease 2017 Study. J Dent Res 2020;99:362-73.
- Guo H, Zhou Y, Liu X, et al. The impact of the COVID-19 epidemic on the utilization of emergency dental services. J Dent Sci 2020;15:564-7. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Dong D, Tang L, Li ZY, et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol 2019;30:431-8. [Crossref] [PubMed]
- Song C, Wang M, Luo Y, et al. Predicting the recurrence risk of pancreatic neuroendocrine neoplasms after radical resection using deep learning radiomics with preoperative computed tomography images. Ann Transl Med 2021;9:833. [Crossref] [PubMed]
- Qian L, Lv Z, Zhang K, et al. Application of deep learning to predict underestimation in ductal carcinoma in situ of the breast with ultrasound. Ann Transl Med 2021;9:295. [Crossref] [PubMed]
- Hwang JJ, Jung YH, Cho BH, et al. An overview of deep learning in the field of dentistry. Imaging Sci Dent 2019;49:1-7. [Crossref] [PubMed]
- Lee JH, Kim DH, Jeong SN, et al. Diagnosis and prediction of periodontally compromised teeth using a deep learning-based convolutional neural network algorithm. J Periodontal Implant Sci 2018;48:114-23. [Crossref] [PubMed]
- Murata S, Lee C, Tanikawa C, et al. Towards a fully automated diagnostic system for orthodontic treatment in dentistry. 2017 IEEE 13th International Conference on e-Science (e-Science); 24-27 October 2017; Auckland, New Zealand. IEEE, 2017.
- Casalegno F, Newton T, Daher R, et al. Caries Detection with Near-Infrared Transillumination Using Deep Learning. J Dent Res 2019;98:1227-33. [Crossref] [PubMed]
- Lee JH, Kim DH, Jeong SN, et al. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent 2018;77:106-11. [Crossref] [PubMed]
- Ding B, Zhang Z, Liang Y, et al. Detection of dental caries in oral photographs taken by mobile phones based on the YOLOv3 algorithm. Ann Transl Med 2021;9:1622. [Crossref] [PubMed]
- Bader JD, Shugars DA, Bonito AJ. Systematic reviews of selected dental caries diagnostic and management methods. J Dent Educ 2001;65:960-8.
- Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari V, Hebert M, Sminchisescu C, et al. editors. Computer Vision – ECCV 2018. Lecture Notes in Computer Science, vol 11211. Springer, Cham, 2018:833-51.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 27-30 June 2016; Las Vegas, NV, USA. IEEE, 2016:770-8.
- Hu J, Shen L, Sun G. Squeeze-and-excitation networks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 18-23 June 2018; Salt Lake City, UT, USA. IEEE, 2018:7132-41.
- Deng J, Dong W, Socher R, et al. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition; 20-25 June 2009; Miami, FL, USA. IEEE, 2009:248-55.
Chen LC Papandreou G Schroff F Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587v3 [cs.CV],2017 .- Milletari F, Navab N, Ahmadi SA. VV-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV); 25-28 October 2016; Stanford, CA, USA. IEEE, 2016:565-71.
- Yu J, Jiang Y, Wang Z, et al. Unitbox: An advanced object detection network. Proceedings of the 24th ACM international conference on Multimedia. 2016:516-20.
- Ando M, González-Cabezas C, Isaacs RL, et al. Evaluation of several techniques for the detection of secondary caries adjacent to amalgam restorations. Caries Res 2004;38:350-6. [Crossref] [PubMed]
- Featherstone JD. The science and practice of caries prevention. J Am Dent Assoc 2000;131:887-99. [Crossref] [PubMed]
- Schwendicke F, Elhennawy K, Paris S, et al. Deep learning for caries lesion detection in near-infrared light transillumination images: A pilot study. J Dent 2020;92:103260. [Crossref] [PubMed]
- Park WJ, Park JB. History and application of artificial neural networks in dentistry. Eur J Dent 2018;12:594-601. [Crossref] [PubMed]
- Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 21-26 July 2017; Honolulu, HI, USA. IEEE, 2017:4700-8.
(English Language Editor: J. Jones)




