
MiR-202-3p inhibits the proliferation and metastasis of lung adenocarcinoma cells by targeting RRM2
Highlight box
Key findings
• MiR-202-3p inhibited the proliferation and metastasis of lung cancer cells by targeting RRM2. LUAD patients with high miR-202-3p or low RRM2 expression had a better prognosis.
What is known and what is new?
• MiR-202-3p was lowly expressed in lung cancer tissue. MiR-202-3p overexpression inhibited the proliferation and metastasis of lung cancer cells. RRM2 was highly expressed in lung cancer tissue and promoted the proliferation and metastasis of lung cancer cells;
• MiR-202-3p targeted and inhibited RRM2, thereby reducing the proliferation and metastasis of LUAD cells. LUAD patients with low RRM2 expression had a better prognosis, and the expression level of RRM2 was correlated with the clinical characteristics of lung cancer patients.
What is the implication, and what should change now?
• Our findings not only further enrich the molecular mechanism of lung cancer development and progression, but also provide a molecular basis for targeted treatment of lung cancer.
Introduction
Lung cancer (LC) has the highest incidence among all malignancies worldwide and is also the most common cause of cancer-related death (1,2). LC can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) based on its histology and etiology. In China, the incidence of NSCLC accounts for approximately 85% of LC cases, and its histology is usually dominated by adenocarcinoma and squamous cell carcinoma (3).
Lung adenocarcinoma (LUAD) is currently the most common histological subtype of primary LC, accounting for approximately 40% of LC cases, and the relative incidence of LUAD is still increasing (4,5). The prognosis of patients with various tissue subtypes of LUAD is very poor (6). Although there have been considerable advancements in treatment strategies for LUAD, the overall short-term survival of NSCLC patients has not exceeded 20% in the past few decades (3). Increasing evidence indicates that malignant proliferation and distant metastasis are the main challenges of cancer treatment (7). Therefore, studying the genes associated with tumor metastasis in the pathophysiological process of LUAD is crucial, and targeting these genes may be a potential approach for the treatment of NSCLC. Although a large number of studies have been carried out on LC, the molecular regulatory mechanisms of the occurrence and development of LC have not been systematically investigated.
MiRNA, the most abundant small RNA, are one of the most abundant gene regulatory molecules and can affect the output of several protein-coding genes in multicellular organisms. They are a class of small non-coding RNAs that are highly conserved in evolution, with a length of approximately 19–22 nucleotides (nt). Mature miRNAs, as posttranscriptional inhibitors, recognize and bind to the 3' untranslated region (3'UTR) of target genes through complete or incomplete complementary pairing, thereby destabilizing target mRNAs and inhibiting the translation of target genes. MiRNAs play important regulatory roles in both animals and plants. An increasing number of studies have shown that miRNAs are extensively involved in basic cellular biological processes, such as cell differentiation, proliferation, growth, migration, and apoptosis, as well as carcinogenic or cancer-inhibiting processes (8-10).
MiR-202-3p is present in many human diseases and is a new potential biomarker for the diagnosis of type 1 gastric neuroendocrine tumors (11). Its overexpression in bone marrow mesenchymal stem cells improves cerebral ischemia-reperfusion injury by promoting angiogenesis and inhibiting inflammation (12). Additionally, the overexpression of nuclear-enriched autosomal transcript 1 promotes the proliferation, migration, and invasion of endometrial cancer cells and inhibits apoptosis by targeting the miRNA-202-3p/T-cell immunoglobulin and mucin domain 4 axis (13). However, the pathophysiological role of miR-202-3p in NSCLC is not completely clear.
Ribonucleotide reductase (RR) is the only rate-limiting enzyme in intracellular DNA synthesis and plays an important role in nucleic acid metabolism. It can catalyze the reduction of nucleoside diphosphate to deoxyribonucleoside diphosphate, which is the rate-limiting step in the biosynthesis of deoxyribonucleoside triphosphate (dNTP) (14). RR includes two subunits, M1 and M2, both of which are dimeric structures. The ribonucleotide reductase regulatory subunit M2 (RRM2) has a site that binds to substrates and allosteric effectors and contains a direct electron-donating group, such as a sulfhydryl, controlling the substrate specificity and enzyme activity and acting as a tumor suppressor gene. RRM2 is an iron-sulfur protein that participates in catalytic reactions through the formation of specific free radicals through the benzene ring of tyrosine residues. RRM2 is not only the catalytic region responsible for substrate conversion but also carries the contact inhibition region and is the main regulatory element of RR activity (15).
RRM2 is involved in the biosynthesis of dNTP and is an indispensable key enzyme in the process of DNA synthesis and repair. Wang et al. used a mouse xenograft model to confirm that RRM2 activates the extracellular regulated protein kinases 1/2 (ERK1/2) pathway through reactive oxygen species (ROS) proto-oncogene 1-receptor tyrosine kinase to promote the formation of blood vessels in cervical cancer cells, increases microvessel density, and promotes the proliferation of tumor cells (16). Research has indicated that RRM2 overexpression increases the invasiveness of pancreatic cancer by enhancing the expression of matrix metallopeptidase 9 (MMP-9) (17). Rasmussen et al. showed that RRM2 affects the upstream tumor suppressor gene BRCA1 to form the BRCA1-RRM2 axis, protecting glioblastoma (GBM) cells from interference by endogenous apoptotic factors such as oncogenes and DNA damage, resulting in GBM invasion and metastasis (18). Wang et al. found that silencing RRM2 inhibits the proliferation, invasion, and migration of GBM cells and increases the rate of apoptosis. RRM2 overexpression promotes cell proliferation, migration, and invasion and inhibits cell apoptosis (19). In vivo, RRM2 overexpression was shown to accelerate the growth of GBM cells; in the study, RRM2 was highly expressed in human GBM cells. RRM2 promotes the proliferation, migration, and invasion of human GBM cells but inhibits cell apoptosis. In recent years, RRM2 has been shown to be overexpressed in a variety of malignant tumors, such as brain GBM, cholangiocarcinoma, pancreatic cancer, breast cancer, NSCLC, ovarian cancer, bladder cancer, and colorectal cancer (20-23). Therefore, studies on the effects of abnormal RRM2 expression on the proliferation, chemotherapy resistance, and metastasis of malignant tumors and the prognosis and survival of patients with such tumors can provide a new theoretical basis for the diagnosis, treatment, and prognosis of clinical malignant tumors. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6089/rc).
Methods
Data
In this study, the mRNA expression data of 594 LUAD transcriptomes were downloaded from The Cancer Genome Atlas (TCGA) database (https://tcga-data.nci.nih.gov/tcga/), and the expression data were sorted and extracted using Perl; a total of 59 normal samples and 535 tumor samples were included. TCGA database was used to obtain the clinical data of 522 LUAD patients, from which survival time, survival status, sex, age, TNM stage, and clinicopathological stage were extracted through Perl. Some clinical characteristic data were incomplete. Cases with unknown data were excluded from the analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
RRM2 survival was analyzed using the survival package in R language. The matrix/immune score was used to divide the samples into high and low groups using the median score as the boundary. A survival curve was plotted using the Kaplan-Meier (K-M) method. The clinicopathological staging data were sorted, and unknown samples were then removed before correlation analysis. Each object in the results was analyzed by paired comparison to obtain the P values.
The differential analysis of gene profiles was performed using the limma package in R language. Differentially expressed genes (DEGs) were screened using |log2(fold change)| >1 and −log10(P value) <0.05 to obtain the upregulated and downregulated gene sets. Next, a volcano plot was constructed. A heatmap was then plotted using the pheatmap package in R language, and 20 upregulated and 20 downregulated genes were presented according to their log2(fold change)| value. Finally, the VennDiagram package in R was used to draw a Venn diagram to obtain the genes related to miR-202-3p in the miRTarBase, MiRDB, and TargetScan databases. These genes and the above DEGs were used to determine the function of the DEGs. To discover the function of the DEGs, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and plotted the enrichment pathway results using the ggplot2 package in R language.
Univariate Cox regression analysis of the clinical prognostic data of RRM2 was performed using the survival package in R language. A Cox risk model was built using R language, and a coefficient was obtained for each gene. The risk score was equal to the sum of the expression level of each gene multiplied by the coefficient, and the model was optimized. First, the survival analysis method described above was used to draw a survival curve of the risk model and a risk curve. The risk model and clinical characteristics and survival prognosis were then subjected to Cox regression analysis, and the results were viewed using forest plots. Next, the survivalROC package in R was used to plot the receiver operating characteristic (ROC) curves for the risk model and clinical characteristics. Finally, the correlation between the risk model and clinical characteristics was analyzed.
Cells and main reagents
A549, H1975, human NSCLC cells, and Beas-2b normal bronchial epithelial cells were purchased from Guangzhou Cellcook Biotechnology Co., Ltd. (China). Fetal bovine serum (FBS), RPMI-1640 medium, and Dulbecco’s Modified Eagle Medium (DMEM) were purchased from Biological Industries (BioInd, Israel). Ham’s F-12K medium was purchased from Gibco (USA), and TRIzol reagent was purchased from Nanjing Vazyme Biotech Co. (China). The miRNA mimic/inhibitor/control were purchased from Guangzhou RibPharm Co., Ltd. (China), trypsin-ethylene diamine tetraacetic acid (EDTA) was purchased from New Cell & Molecular Biotech Co., Ltd. (Suzhou, China), JetPrime transfection reagent was purchased from Polyplus Co., Ltd. (France), and ChamQ Universal SYBR quantitative polymerase chain reaction(qPCR) Master Mix reagent and HiScript III All-in-one RT SuperMix Perfect for qPCR were purchased from Nanjing Vazyme Biotech Co., Ltd. The RRM2 (ab57653) and β-actin (ab8226) antibodies were purchased from Abcam (USA), and the goat anti-mouse secondary antibody and goat anti-rabbit secondary antibody were purchased from Shanghai Beyotime Biotechnology (China). The Cell Counting Kit-8 (CCK-8) reagent was purchased from Tongren Company (Japan), and small interfering RNA (siRNA) was purchased from Suzhou GenePharma Co., Ltd. (China).
Cell culture
The A549 and H1975 human LUAD cell lines and human normal bronchial epithelial cell line Beas-2b were selected for culture. A549 cells were cultured in F-12k medium, H1975 cells were cultured in RPMI-1640 medium, and Beas-2b cells were cultured in DMEM. The above medium was supplemented with 10% FBS and 1% penicillin-streptomycin. The cells were cultured in a 37 ℃ incubator with 5% carbon dioxide (CO2). The medium was changed every other day, and the original medium was discarded when changing the medium. The plates were rinsed with phosphate-buffered saline (PBS) three times; to avoid contamination, all PBS was aspirated after each rinse, after which fresh medium was added. The growth of the cells was observed using a microscope, and a growth curve was plotted. When the cells were in the logarithmic growth phase, 0.5–1 mL of trypsin digestion solution containing 0.25% EDTA was added, and the cells were placed in a 37 ℃, 5% CO2 incubator for 2 min.
The state of the cells was observed under a microscope (NIKON, Japanese). When the cells became round with an increase in intercellular gaps, 3 mL of the complete medium containing 10% FBS was added to terminate the digestion, and the cells that were still adherent were pipetted into the cell suspension. Next, the cell suspension was placed in a centrifuge tube and centrifuged at 1,000 rpm for 5 min. The supernatant was then discarded, the cell pellet was resuspended, and a new medium containing 10% FBS was added. The cells were passaged every 2–3 days. Cells in the logarithmic growth phase were used in all experiments.
Cell transfection
Oligonucleotides were synthesized by RiboBio Co., Ltd. (RiboBio, Guangzhou, China). SiRNA was provided by GenePharma (Suzhou, China). The cells were divided into a scrambled group, a miR-202-3p mock group, a miR-202-3p inhibitor group, a si-RRM2 negative control (NC) group, a miR-202-3p inhibitor + RRM2 siRNA group, and a RRM2 siRNA group. Cells in the logarithmic growth phase (2.0×106 cells/well) were cultured in six-well plates without antibiotics for 24 h before transfection. When the cells were 80% confluent, 50 nM miR-202-3p NC, 20 nM miR-202-3p mimics, 100 nM miR-202-3p inhibitor, 100 nM si-NC, and 100 nM miR-202-3p inhibitor + RRM2 siRNA or 100 nM RRM2 siRNA were transfected using polyplus JetPrime. Forty-eight hours after transfection, the cells were collected for subsequent experiments.
Analysis of dual luciferase reporter genes
The Promega Dual-Luciferase system 1.5× passive lysis buffer (PLB) was diluted with distilled water to 1× PLB, 100 µL of PBS was added to each well of a 96-well plate, and the cells were dispersed by pipetting. The plates were placed on a shaker at room temperature. After shaking for 15 min, the cell lysate was aspirated into a 1.5 mL centrifuge tube, and the lysate was centrifuged at 12,000 rpm at 4 ℃ for 10 min. The supernatant was then transferred to a new tube.
Next, 100 µL of Luciferase Assay Reagent II [Luciferase Assay Reagent, Promega (Madison, WI, USA)] was added to each well of the 96-well plate and 20 µL of cell lysate was also added, followed by mixing by pipetting 2–3 times. The firefly luciferase value was then measured and recorded, serving as the internal reference value. Subsequently, 100 µL of Stop & Glo® Reagent (Luciferase Assay Reagent, Promega) was added, followed by mixing by pipetting 2–3 times. The Renilla luciferase value was then measured and recorded, serving as the luminescence value for the reporter gene.
Transwell migration experiment
Cell migration experiments were performed in 24-well plates (Corning, New York, USA). A549 cells (2.5×105 cells/well) were seeded into the culture medium without FBS in the upper chamber, and a complete culture medium containing 10% FBS was added to the lower chamber. After 24 h, the cells that migrated to the lower surface of the filter were fixed with 4% paraformaldehyde (PFA) and incubated with 4',6-Diamidino-2'-phenylindole (DAPI, 1:1,000, Invitrogen, CA, USA) at room temperature for 5 min. Migrated cells were counted in five random areas under an inverted fluorescence microscope (Leica DMI6000B, Wetzlar, Germany). All experiments were performed in triplicate.
Real-time polymerase chain reaction (RT-PCR)
Reverse transcription was performed using HiScript III RT SuperMix for qPCR (Nanjing Vazyme Biotech Co., Ltd.). RT-PCR was performed using the ChamQ Universal SYBR qPCR Master Mix kit (Nanjing Vazyme Biotech Co., Ltd.) and a Thermo Fisher StepOne (USA) RT-PCR system. U6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as internal controls. The reverse primer sequence for U6 was 5'-AACGCTTCACGAATTTGCGT-3', and the forward primer sequence was 5'-CTCGCTTCGGCAGCACA-3'; the reverse primer sequence for GAPDH was 5'-GGAGCGAGATCCCTCCAAAAT-3', and the forward primer sequence was 5'-GGCTGTTGTCATACTTCTCATGG-3'; the forward primer sequence for RRM2 was 5'-TTGAAACGATGCCTTGTGTCA-3', and the reverse sequence was 5'-ACCATAGGTAGCCTCTTTGTCC-3'; and the primers for miR-202-3p were Bulge-loopTM miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific to miR-202-3p and designed by RiboBio (Guangzhou, China). All experiments were repeated three times. The qPCR conditions for RRM2 were as follows: predenaturation at 95 ℃ for 30 s; amplification (40 cycles) at 95 ℃ for 10 s and 60 ℃ for 30 s; and 95 ℃ for 15 s, 60 ℃ for 60 s, and 95 ℃ for 15 s. The relative expression level of the target gene was calculated using the 2−ΔΔCt method. The qPCR conditions for miR-202-3p were as follows: predenaturation at 95 ℃; amplification at 95 ℃ for 2 s, 60 ℃ for 20 s, and 70 ℃ for 10 s; and 95 ℃ for 15 s, 60 ℃ for 60 s, and 95 ℃ for 15 s. The relative expression level of the target gene was calculated using the 2−ΔΔCt method.
CCK-8 test
CCK-8 (Dojindo, Kumamoto, Japan) was used according to the manufacturer’s instructions. A549 cells were seeded in 96-well plates (3×103 cells/well) and cultured overnight. The absorbance at 450 nm was measured using a microplate reader (Shanghai, Thermo Fisher Science, Shanghai, China) for 4 consecutive days. This experiment was repeated three times independently.
Western blot (immunoblot)
Cells were collected and lysed in Radio-Immunoprecipitation Assay (RIPA) lysis buffer containing protease inhibitors. After centrifugation, the supernatant was mixed with dithiothreitol (DTT)/2× sodium dodecyl sulfate (SDS) loading buffer and boiled in water. Then total proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membrane was immersed in a blocking solution [5% bovine serum albumin (BSA) dissolved in PBST (PBS pH 7.5, 0.1% Tween 20)] for 1 h at room temperature. The membrane was washed with PBST, incubated with primary antibody [rabbit anti-human RRM2 antibody (1:1,000) and mouse anti-human β-actin antibody (1:1,000)] at 4 ℃ overnight, and washed again with PBST for three times. The membranes were incubated at room temperature with the appropriate secondary antibodies [horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:1,000) and HRP-conjugated goat anti-mouse secondary antibody (1:1,000)] for 1 h and rinsed with PBST for three times. Chemiluminescence detection system (Bio Rad Laboratories, Hercules, USA) was used to detect gray stripe. Image J software was used to analyze the grayscale of the strips.
Statistical analysis
In this study, R language was used to perform the statistical analyses. The Wilcoxon rank sum test was employed to screen the genes. Significance tests were applied for the correlations. One-way analysis of variance and t-tests were utilized to detect differences. Univariate and multivariate Cox analyses and the construction of the Cox models were performed using R language. All experiments were repeated three times independently, and the data were expressed as the mean ± standard deviation (SD). The independent sample t-test was used to compare the means of unpaired groups. The figures were created in R language and GraphPad Prism 8 (GraphPad Software, San Diego, CA).
Results
Expression of miR-202-3p in various tumor tissues and ROC analysis
The expression level of miR-202-3p in various tumors was analyzed using CancerMIRNome. The expression of miR-202-3p in LUAD was low (P<0.05) (Figure 1A), and the area under the ROC curve (AUC) for LUAD was 0.77 (P<0.05). This finding indicates that miR-202-3p has a high sensitivity in the diagnosis of LUAD (Figure 1B,1C).
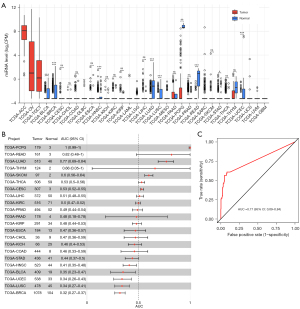
Changes in miR-202-3p expression in NSCLC
The analysis of CancerMIRNome indicated that the expression of miR-202-3p was low in LUAD tissue, P<0.05 (Figure 2A), which was also confirmed by qPCR in Beas-2b, A549, and H1975 cells, i.e., the expression of miR-202-3p was higher in the normal epithelial cells than in the LC cell lines (Figure 2B).
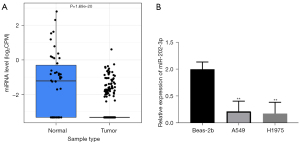
Detection of miR-202-3p expression in A549 cells after transfection
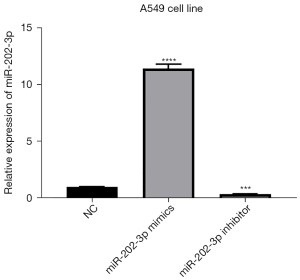
The miR-202-3p mimics, miR-202-3p inhibitor, and miR-NC were transfected into A549 cells. Forty-eight hours after transfection, the expression level of miR-202-3p was detected by qPCR. The results showed that compared with that in the miR-NC group, the expression level of miR-202-3p in the mimic group was significantly higher than that in the miR-NC group and that in the inhibitor group was lower than that in the miR-NC group (P<0.05, Figure 3).
MiR-202-3p inhibits the proliferation of LUAD cells
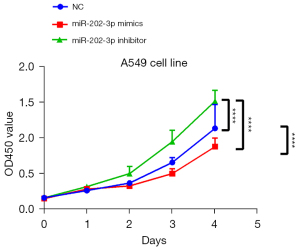
The effects of miR-202-3p and RRM2 on the proliferation of A549 cells were examined using CCK-8 assays. The results showed that the growth rate of the miR-202-3p mimics group was lower than that in the NC group 48 h after transfection. In contrast, the growth rate of the miR-202-3p inhibitor group was significantly higher than that of the NC group (Figure 4), indicating that miR-202-3p inhibits the proliferation of LUAD cells.
MiR-202-3p inhibits the migration of LUAD cells
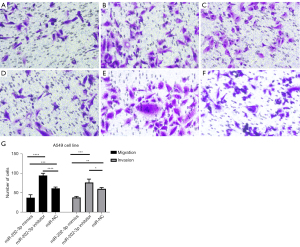
The effect of miR-202-3p on the migration ability of A549 cells was detected using the Transwell method. The results of the migration and invasion experiments indicated that the miR-202-3p inhibitor group had the most cells pass through the Transwell membrane, followed by the miR-202-3p NC group and the miR-202-3p mimics group, which had the fewest cells pass through the membrane (Figure 5). Therefore, it can be concluded that miR-202-3p inhibits the migration and invasion of LUAD cells.
Prediction of miR-202-3p-related genes
We performed bioinformatics analysis of NSCLC using TCGA, which is a public gene expression dataset. Next, the DEGs between the tumor and normal samples were identified using the Wilcoxon rank sum test. The DEGs associated with miR-202-3p were identified and divided into high- and low-expression groups (Figure 6A). Cluster analysis of the DEGs revealed 20 upregulated genes and 20 downregulated genes (Figure 6B). A comparison of the three databases (i.e., mRDB, miRTarBase, and TargetScan) showed that among the predicted genes related to miR-202-3p, 75 genes intersected in the three databases (Figure 6C). Moreover, comparing the target mRNAs and the upregulated DEGs, 29 genes intersected (Figure 6D).
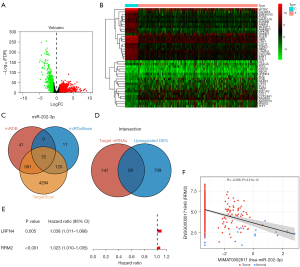
Subsequent univariate Cox proportional hazard regression analysis of these genes showed that RRM2 and leucine rich repeat and fibronectin type III domain containing 4 (LRFN4) were related to clinical prognosis (Figure 6E). Based on a previous study on RRM2 in LC, the role of RRM2 in LC is more important than that of LRFN4 (24). Thus, the RRM2 gene was selected for further study. Analysis of the CancerMIRNome database showed that miR-202-3p was negatively correlated with RRM2 expression, with a correlation coefficient of R=−0.296, P<0.05 (Figure 6F).
Expression of RRM2 in LC cells
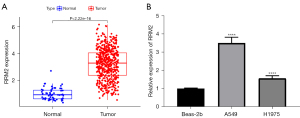
TCGA database also showed that RRM2 was highly expressed in LC tissue (Figure 7A). qPCR confirmed that the relative expression of RRM2 in Beas-2b normal tracheal epithelial cells was lower than that in the LC cell lines (Figure 7B).
MiR-202-3p directly targets RRM2
We also constructed a luciferase reporter vector to analyze dual luciferase reporter genes. The binding of miR-202-3p to the h-RRM2-3UTR is shown in Figure 8A. The dual luciferase reporter gene analysis of H293T cells confirmed that RRM2 contains a direct binding site for miR-202-3p. As shown in Figure 8B, following transfection of the miR-202-3p mimics, the luciferase activity in the h-RRM2-3UTR-wild type (wt) group was significantly decreased, while that in the h-RRM2-3UTR-mutant (mu) group did not change. These results confirmed that miR-202-3p directly targets the RRM2 gene.
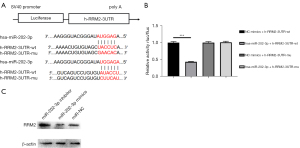
Western blot assays were used to detect the expression of the RRM2 protein after transfection, and the results are shown in Figure 8C. Compared with that in the miR-NC group, the expression of the RRM2 protein was higher in the miR-202-3p inhibitor group, and the expression of the RRM2 protein in the miR-202-3p mimics group was markedly lower; the difference was statistically significant (P<0.05).
Clinical correlation between RRM2 expression and LUAD
To better understand whether RRM2 expression is related to the occurrence and development of LC, the correlation between RRM2 expression and clinicopathological stage was further analyzed. From the perspective of tumor size and depth of invasion (T), the expression level of RRM2 was the lowest in stage T1. Although the expression level of RRM2 in stages T2 to T4 was T4 > T2 > T3, only the expression levels of RRM2 in T1 and T2 were correlated with T stage (P<0.05); there were no other significant correlations (Figure 9A). From the perspective of lymph node metastasis (N), although N3 > N2 > N1 > N0, only N0 and N1 and N0 and N2 were statistically correlated (P<0.05, Figure 9B). In terms of distant metastasis (M), the level of RRM2 expression was higher for M1 than for M0 metastasis, i.e., the level of RRM2 expression was correlated with distant metastasis (P=0.069), but this was not statistically significant (Figure 9C). Finally, for clinical stage, the level of RRM2 expression was positively correlated with stage, and there was a significant difference between stage I and stages II, III, and IV (P<0.05, Figure 9D). The above data indicate that the level of RRM2 expression is strongly correlated with TNM staging.
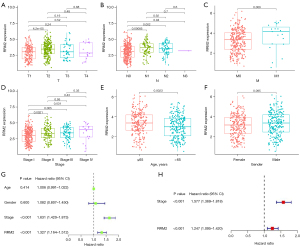
The correlation between RRM2 expression level and age (>65 and ≤65 years) and sex was further analyzed. The results showed that the level of RRM2 expression in patients aged ≤65 years was higher than that in patients aged >65 years, P<0.05 (Figure 9E). Also, the level of RRM2 expression in males was higher than that in females, with a P value of 0.095, i.e., not significant (Figure 9F). Subsequently, univariate and multivariate Cox regression analyses were performed to further evaluate the performance of our model in LUAD patients and using other common prognostic factors. The univariate Cox analysis results showed that clinicopathological stage was closely related to the clinical survival prognosis of LUAD patients (Figure 9G). When these factors were included in the multivariate Cox analysis, only clinicopathological stage could be used as an independent risk factor for the prognosis of LUAD patients (Figure 9H).
Analysis of RRM2 expression and survival prognosis and ROC curve of LC patients
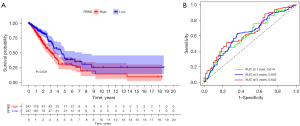
To explore the correlation between RRM2 expression and survival, survival analysis was performed using RNA-seq data for LUAD in TCGA database and clinical data for LUAD patients downloaded from the UCSC (University of California Santa Cruz) XENA database. The K-M survival curve showed that the survival rate of the RRM2 high-expression group was lower than that of the RRM2 low-expression group (P<0.05, Figure 10A). To verify the diagnostic value of RRM2, we generated a ROC curve for patient RRM2 expression levels; the AUCs at 1, 3, and 5 years were 0.614, 0626, and 0.642, respectively (Figure 10B), indicating that RRM2 has a strong diagnostic effect in LC.
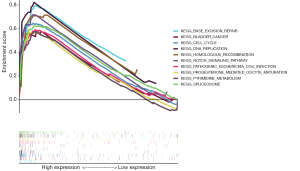
KEGG enrichment analysis of RRM2
To further understand the role of the RRM2 protein in cellular metabolic activities and the molecular functions, biological processes, and signaling pathways involved, KEGG pathway enrichment analysis was performed for RRM2 using the clusterProfiler software package in R language. KEGG analysis revealed RRM2 enrichment in the Notch pathway (Figure 11).
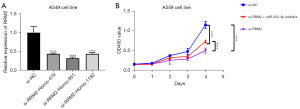
RRM2 promotes the proliferation of LUAD cells
Following transfection of A549 cells with si-RRM2-Homo-479, si-RRM2-Homo-951, and si-RRM2-Homo-1182, the expression of RRM2 in the above three siRNA groups was lower than that in the si-NC group (P<0.05, Figure 12A). Si-RRM2-Homo-951 led to the greatest decrease in RRM2 and was thus used in subsequent experiments. The growth rate of A549 cells in the si-RRM2 group was slower than that of A549 cells in the si-NC group and the si-RRM2 + miR-202-3p inhibitor group. Also, the growth rate of A549 cells in the si-RRM2 + miR-202-3p inhibitor group was slower than that of A549 cells in the si-NC group but faster than that of A549 cells in the si-RRM2 group, indicating that RRM2 promoted the proliferation of A549 cells and that the miR-202-3p inhibitor reduced the inhibitory effect of si-RRM2 on the growth of A549 cells (Figure 12B).
RRM2 promotes the migration and invasion of LUAD cells
Transwell assays were applied to assess the migration and invasion of LUAD cells in the si-RRM2 group, the miR-202-3p inhibitor + si-RRM2 group, and the si-NC group. The si-NC group had the most cells pass through the Transwell, followed by the miR-202-3p inhibitor + si-RRM2 group and the si-RRM2 group, which had the fewest number of cells pass through the Transwell. All P values were less than 0.05 in the intergroup comparisons (Figure 13). This finding indicates that RRM2 promotes the invasion and metastasis of A549 cells.
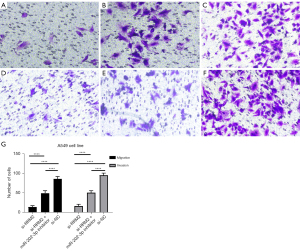
Previous experiments have shown that miR-202-3p directly targets RRM2; furthermore, the number of migrating and invading cells in the miR-202-3p inhibitor group was higher than that in the si-RRM2 group but less than the si-NC group, indicating that the miR-202-3p inhibitor alleviated the inhibitory effect of si-RRM2 on RRM2, further demonstrating the direct targeting effect of miR-202-3p on RRM2.
Discussion
In recent decades, the incidence of LC has increased significantly due to air pollution and other factors, of which the increase in the incidence of NSCLC has been the most significant (25,26). Although a large proportion of NSCLC patients are diagnosed early and receive treatment, the overall prognosis of LC patients is very poor due to frequent recurrence and metastasis (27).
MiRNAs are small non-coding RNAs that act as posttranscriptional mediators of gene expression through the silencing or degradation of mRNAs (messenger RNAs). In recent years, numerous studies have been conducted on miRNAs, and there is a considerable amount of evidence showing that there are many connections between miRNAs and the occurrence and development of tumors. The occurrence and development of several cancers are closely related to the dysregulation of miRNAs. MiRNAs act as tumor suppressors or oncogenes (oncomiRs), and miRNA mimics and miRNA targeting molecules (antimiRs) have shown promise in preclinical development (28). Recently, a previous study has demonstrated that miRNAs can regulate the development of a variety of tumors and have important effects on tumor development, prognosis, and treatment (29). MiRNAs inhibit different target genes to affect the proliferation, migration, and invasion of cancer cells. For example, the overexpression of miR-25-3p inhibits regulator of G-protein signaling (RGS3). Also, the inhibition of miR-25 promotes the apoptosis and delays the proliferation of NSCLC cells (30). MiR-125b can promote tumor metastasis in human NSCLC cells by targeting tumor protein 53-induced nuclear protein 1 (TP53INP1) (31). Additionally, miR-31 overexpression may be associated with cisplatin (DDP) resistance, and miR-31 can exert an anti-apoptotic effect by inhibiting the 3'UTR of ABC transporter 9 protein (ABCB9), which is a key factor in the mediation of DDP resistance (32). MiR-31 may also promote the development of LC by stimulating mutant Kirsten rat sarcoma viral oncogene homolog (KRAS) (33). In summary, the identification of miRNAs and their target genes and pathways associated with NSCLC can be used as an entry point to develop treatments for LC, which is a highly malignant tumor in clinical practice.
There is evidence that miR-202-3p is associated with a variety of human diseases. For example, the silencing of miR-202-3p in metastatic breast cancer can promote the expression of matrix metalloproteinase 1 (MMP-1) and an infiltrating phenotype in brain cells (34); miR-202-3p can target brain-derived neurotrophic factors and participate in depression-like behaviors (35). MiRNA-202-3p can also target transient receptor potential cation channel, subfamily M, member 6 (TRPM6) and protect the heart from myocardial ischemia-reperfusion injury by activating TGF-cads1/Smad signaling (36). However, the mechanism by which miR-202-3p promotes LC metastasis and proliferation in NSCLC is still unknown.
In this study, the expression of miR-203-3p in NSCLC tissue and cells was significantly lower than that in adjacent tissue and normal lung cells. Subsequently, we validated this hypothesis using data analysis and qPCR experiments. We focused on elucidating the potential mechanism of miR-202-3p-mediated promotion of LUAD metastasis. We observed that the expression level of miR-202-3p was significantly decreased in NSCLC tissue and cells, while its overexpression inhibited the proliferation and migration of tumor cells in vitro. The TargetScan data analysis conducted in this study also showed that RRM2 might directly interact with miR-202-3p. In other words, miR-202-3p might target and regulate RRM2. To verify this hypothesis, we performed a dual luciferase reporter gene analysis, and the results indicated that miR-202-3p could directly target RRM2.
The full name of RRM2 is ribonucleoside reductase M2, which is a subunit of RR. RR is the only enzyme that can catalyze the de novo formation of deoxyribonucleoside and is critical in DNA synthesis. High RRM2 expression is very common in cancer, and RRM2 is considered to be a promoting factor and therapeutic target for the occurrence and development of tumors (37). Numerous studies have shown that RRM2 expression is related to the activity of chemotherapeutic drugs in cancer (37-39). High RRM2 expression is associated with low sensitivity to chemotherapy and poor prognosis (40). Using multi-omics and bioinformatics analysis, RRM2 upregulation was shown to be associated with chemotherapy resistance and poor prognosis; however, these results need to be verified in in vivo and in vitro experiments and multicenter randomized controlled clinical trials (41). The upregulation of RRM2 affects complex signaling pathways and mediates the survival, proliferation, migration, invasion, and chemotherapy resistance of cancer cells. Therefore, RRM2 may be a potential target for overcoming chemotherapy resistance. RRM2 not only has broad application prospects in chemotherapy but also has clinical significance in terms of cancer diagnosis and the prediction of patient prognosis, as verified by numerous studies. For example, high RRM2 expression in LC can be used as an independent predictor of poor prognosis (42).
In terms of mechanism, miR-202-3p inhibited RRM2-mediated proliferation and invasion of NSCLC cells by inhibiting RRM2. These results suggest that miR-202-3p may be a therapeutic molecule for the clinical treatment of NSCLC, a dangerous malignant tumor.
Through data analysis carried out in this study, we found that RRM2 expression was low in paracancerous tissue and high in cancer tissue. An analysis of TCGA data and qPCR of normal lung epithelial cells and lung SCLC cells confirmed the high expression of RRM2 in LC tissue. Moreover, the CCK-8 and Transwell assays performed in this study demonstrated that RRM2 promoted the proliferation and metastasis of LC cells. In the co-transfection experiment with a miR-202-3p inhibitor and si-RRM2, the results showed that the proliferation and metastasis of cells in the transfection group were faster than those of cells in the si-RRM2 group but slower than those of cells in the si-NC group, confirming that miR-202-3p and RRM2 have a targeted effect. High RRM2 expression is common in cancer.
We also found that the overexpression of miR-202-3p inhibited the proliferation and metastasis of LUAD. MiRNAs play a gene regulatory role by binding to the 3'UTR of target mRNA. We identified the enrichment of RRM2 in the Notch pathway. Notch signaling regulates a variety of cell fate decisions in multiple tissues, including lineage commitment, differentiation, cell cycle progression, as well as stem cell maintenance and self-renewal (43,44). Studies on the role of Notch signaling in tumorigenesis have mainly focused on the role of Notch as an oncogene; Notch is considered to be an oncogene in several cancers, including brain cancer, breast cancer, ovarian cancer, and NSCLC (45-47). These findings can provide some ideas for further research on RRM2. In recent years, gene transfection has been widely used to regulate gene expression. These findings can provide new ideas and directions for the treatment of LUAD.
In summary, our study showed that the expression of miR-202-3p was downregulated in NSCLC tumor tissue and LUAD cells and that the expression of RRM2 was upregulated. In addition, we also demonstrated the direct-targeted regulation of miR-202-3p and RRM2 and demonstrated that miR-202-3p inhibited the proliferation and metastasis of LUAD through the targeted inhibition of RRM2. Furthermore, there was a correlation between the RRM2 expression level and clinical characteristics, demonstrating that RRM2 has a certain value in predicting the prognosis of and diagnosing LUAD. These findings provide effective molecular targets for the treatment of LUAD.
Conclusions
In this study, LUAD RNA-seq data from TCGA were used as subjects, and the genes targeted by miR-202-3p were identified through serial analysis. Next, miR-202-3p and the targeted gene, RRM2, were validated. Furthermore, the roles of miR-202-3p and RRM2 in LC cells were explored. The pathology and prognosis of RRM2 in LC patients were also evaluated through data analysis. The main conclusions were as follows: (I) msiR-202-3p is lowly expressed in LC tissue, and miR-202-3p overexpression inhibits the proliferation and metastasis of LC cells; (II) RRM2 is highly expressed in LC tissue and promotes the proliferation and metastasis of LC cells; (III) miR-202-3p inhibits RRM2, thereby reducing the proliferation and metastasis of LUAD cells; and (IV) the prognosis of LUAD patients with low RRM2 expression is good, and the level of RRM2 expression is correlated with the clinical characteristics of LC patients.
Acknowledgments
Funding: This work was supported by Natural Science Research Project of Nantong Science and Technology Bureau (No. MS12020029).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6089/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6089/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6089/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Liu D. CAR-T "the living drugs", immune checkpoint inhibitors, and precision medicine: a new era of cancer therapy. J Hematol Oncol 2019;12:113. [Crossref] [PubMed]
- Wang S, Zimmermann S, Parikh K, et al. Current Diagnosis and Management of Small-Cell Lung Cancer. Mayo Clin Proc 2019;94:1599-622. [Crossref] [PubMed]
- Meza R, Meernik C, Jeon J, et al. Lung cancer incidence trends by gender, race and histology in the United States, 1973-2010. PLoS One 2015;10:e0121323. [Crossref] [PubMed]
- Zhang C, Yin K, Liu SY, et al. Multiomics analysis reveals a distinct response mechanism in multiple primary lung adenocarcinoma after neoadjuvant immunotherapy. J Immunother Cancer 2021;9:e002312. [Crossref] [PubMed]
- Noma D, Inamura K, Matsuura Y, et al. ALK-rearranged lung adenocarcinoma showing intra-bronchial protrusion: a case of actually peripheral origin with a rare spreading pattern. Ann Clin Oncol 2017;1:15.
- Zhao Y, Brasier AR. Uronic acid pathway metabolites regulate mesenchymal transition and invasiveness in lung adenocarcinoma. Biotarget 2019;3:19. [Crossref] [PubMed]
- Kabekkodu SP, Shukla V, Varghese VK, et al. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc 2018;93:1955-86. [Crossref] [PubMed]
- Ali Syeda Z, Langden SSS, Munkhzul C, et al. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci 2020;21:1723. [Crossref] [PubMed]
- Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech 2021;14:dmm047662. [Crossref] [PubMed]
- Dou D, Li XK, Xia QS, et al. Circulating miRNA-202-3p is a potential novel biomarker for diagnosis of type 1 gastric neuroendocrine neoplasms. BMC Gastroenterol 2021;21:188. [Crossref] [PubMed]
- Yu G, Sun W, Wang W, et al. Overexpression of microRNA-202-3p in bone marrow mesenchymal stem cells improves cerebral ischemia-reperfusion injury by promoting angiogenesis and inhibiting inflammation. Aging (Albany NY) 2021;13:11877-88. [Crossref] [PubMed]
- Xu C, Zhai J, Fu Y. Overexpression of Nuclear Enriched Autosomal Transcript 1 Facilitates Cell Proliferation, Migration Invasion, and Suppresses Apoptosis in Endometrial Cancer by Targeting MicroRNA-202-3p/T Cell Immunoglobulin and Mucin Domain 4 Axis. Cancer Biother Radiopharm 2022;37:815-23. [Crossref] [PubMed]
- Lamb NA, Bard JE, Loll-Krippleber R, et al. Complex mutation profiles in mismatch repair and ribonucleotide reductase mutants reveal novel repair substrate specificity of MutS homolog (MSH) complexes. Genetics 2022;221:iyac092. [Crossref] [PubMed]
- Wu Z, Zhan Y, Wang L, et al. Identification of osalmid metabolic profile and active metabolites with anti-tumor activity in human hepatocellular carcinoma cells. Biomed Pharmacother 2020;130:110556. [Crossref] [PubMed]
- Wang N, Zhan T, Ke T, et al. Increased expression of RRM2 by human papillomavirus E7 oncoprotein promotes angiogenesis in cervical cancer. Br J Cancer 2014;110:1034-44. [Crossref] [PubMed]
- Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun 2007;354:190-6. [Crossref] [PubMed]
- Rasmussen RD, Gajjar MK, Tuckova L, et al. Author Correction: BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun 2018;9:5396. [Crossref] [PubMed]
- Wang M, Chen X, Jin W, et al. Ginsenoside Rb3 exerts protective properties against cigarette smoke extract-induced cell injury by inhibiting the p38 MAPK/NF-κB and TGF-β1/VEGF pathways in fibroblasts and epithelial cells. Biomed Pharmacother 2018;108:1751-8. [Crossref] [PubMed]
- Rahman MA, Amin AR, Wang D, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res 2013;19:3416-28. [Crossref] [PubMed]
- Xia G, Wang H, Song Z, et al. Gambogic acid sensitizes gemcitabine efficacy in pancreatic cancer by reducing the expression of ribonucleotide reductase subunit-M2 (RRM2). J Exp Clin Cancer Res 2017;36:107. [Crossref] [PubMed]
- Kałuzińska Ż, Kołat D, Bednarek AK, et al. PLEK2, RRM2, GCSH: A Novel WWOX-Dependent Biomarker Triad of Glioblastoma at the Crossroads of Cytoskeleton Reorganization and Metabolism Alterations. Cancers (Basel) 2021;13:2955. [Crossref] [PubMed]
- Liu Q, Guo L, Qi H, et al. A MYBL2 complex for RRM2 transactivation and the synthetic effect of MYBL2 knockdown with WEE1 inhibition against colorectal cancer. Cell Death Dis 2021;12:683. [Crossref] [PubMed]
- Ma C, Luo H, Cao J, et al. Independent prognostic implications of RRM2 in lung adenocarcinoma. J Cancer 2020;11:7009-22. [Crossref] [PubMed]
- Urman A, Dean Hosgood H. Lung Cancer Risk, Genetic Variation, and Air Pollution. EBioMedicine 2015;2:491-2. [Crossref] [PubMed]
- Ganti AK, Klein AB, Cotarla I, et al. Update of Incidence, Prevalence, Survival, and Initial Treatment in Patients With Non-Small Cell Lung Cancer in the US. JAMA Oncol 2021;7:1824-32. [Crossref] [PubMed]
- Schanne DH, Heitmann J, Guckenberger M, et al. Evolution of treatment strategies for oligometastatic NSCLC patients - A systematic review of the literature. Cancer Treat Rev 2019;80:101892. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. [Crossref] [PubMed]
- Chen Z, Wu Y, Meng Q, et al. Elevated microRNA-25 inhibits cell apoptosis in lung cancer by targeting RGS3. In Vitro Cell Dev Biol Anim 2016;52:62-7. [Crossref] [PubMed]
- Li Q, Han Y, Wang C, et al. MicroRNA-125b promotes tumor metastasis through targeting tumor protein 53-induced nuclear protein 1 in patients with non-small-cell lung cancer. Cancer Cell Int 2015;15:84. [Crossref] [PubMed]
- Dong Z, Zhong Z, Yang L, et al. MicroRNA-31 inhibits cisplatin-induced apoptosis in non-small cell lung cancer cells by regulating the drug transporter ABCB9. Cancer Lett 2014;343:249-57. [Crossref] [PubMed]
- Edmonds MD, Boyd KL, Moyo T, et al. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J Clin Invest 2016;126:349-64. [Crossref] [PubMed]
- Harati R, Hafezi S, Mabondzo A, et al. Silencing miR-202-3p increases MMP-1 and promotes a brain invasive phenotype in metastatic breast cancer cells. PLoS One 2020;15:e0239292. [Crossref] [PubMed]
- Zhou B, Zhu H, Luo H, et al. MicroRNA-202-3p regulates scleroderma fibrosis by targeting matrix metalloproteinase 1. Biomed Pharmacother 2017;87:412-8. [Crossref] [PubMed]
- Wu HY, Wu JL, Ni ZL. Overexpression of microRNA-202-3p protects against myocardial ischemia-reperfusion injury through activation of TGF-β1/Smads signaling pathway by targeting TRPM6. Cell Cycle 2019;18:621-37. [Crossref] [PubMed]
- Aye Y, Li M, Long MJ, et al. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 2015;34:2011-21. [Crossref] [PubMed]
- Ding J, Li F, Cong Y, et al. Trichostatin A inhibits skeletal muscle atrophy induced by cigarette smoke exposure in mice. Life Sci 2019;235:116800. [Crossref] [PubMed]
- Zhou S, Li J, Xu H, et al. Liposomal curcumin alters chemosensitivity of breast cancer cells to Adriamycin via regulating microRNA expression. Gene 2017;622:1-12. [Crossref] [PubMed]
- Mao G, Li L, Shan C, et al. High expression of RRM2 mediated by non-coding RNAs correlates with poor prognosis and tumor immune infiltration of hepatocellular carcinoma. Front Med (Lausanne) 2022;9:833301. [Crossref] [PubMed]
- Liu R, Guo CX, Zhou HH. Network-based approach to identify prognostic biomarkers for estrogen receptor-positive breast cancer treatment with tamoxifen. Cancer Biol Ther 2015;16:317-24. [Crossref] [PubMed]
- Jin CY, Du L, Nuerlan AH, et al. High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging (Albany NY) 2020;13:3518-35. [Crossref] [PubMed]
- Cai H, Guo H, Deng Y, et al. RRM2 regulates osteogenesis of mouse embryo fibroblasts via the Wnt/β-catenin signaling pathway. Exp Ther Med 2022;24:605. [Crossref] [PubMed]
- Li C, Zheng J, Chen S, et al. RRM2 promotes the progression of human glioblastoma. J Cell Physiol 2018;233:6759-67. [Crossref] [PubMed]
- Teodorczyk M, Schmidt MHH. Notching on Cancer's Door: Notch Signaling in Brain Tumors. Front Oncol 2014;4:341. [Crossref] [PubMed]
- Zhou B, Lin W, Long Y, et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct Target Ther 2022;7:95. [Crossref] [PubMed]
- Ferreira A, Aster JC. Notch signaling in cancer: Complexity and challenges on the path to clinical translation. Semin Cancer Biol 2022;85:95-106. [Crossref] [PubMed]


