
Analyzing the multi-target pharmacological mechanism of folium Artemisia argyi acting on breast cancer: a network pharmacology approach
Highlight box
Key findings
• We explore the pharmacological mechanisms of FAA for breast cancer by a network pharmacology approach.
What is known and what is new?
• Many studies have verified that FAA exerts remarkable antitumor functions.
• The quercetin served as the major ingredient of FAA and might exert its anti-tumor effect mainly by acting on AKT pathway in breast cancer. The anti-tumor mechanism of FAA might be attributed to the coordinated regulation of several cancer-related pathways.
What is the implication, and what should change now?
• It provides a new theoretical basis and some new ideas for the studies of the treatment of breast cancer. It provides the feasibility of experimental research to study the mechanism of FAA in breast cancer, and then provides the possibility to find a new treatment strategy of breast cancer.
Introduction
Breast cancer is the most frequent malignancy occurring in women with 1.38 million new cases each year and nearly 0.46 million related deaths globally (1,2). According to current projections, there will be approximately 3.2 million new cases per year by 2050 (3). Meanwhile, breast cancer is a kind of heterogeneous disease, with differences in occurrence, development, treatment, and prognosis (4). At present, comprehensive adjuvant treatments containing chemotherapy, radiotherapy, endocrine, and HER2-targeted therapies are widely used according to the five major molecular subtypes of breast cancer (5). However, these treatments are costly and usually result in a series of short- and long-term side effects, such as febrile neutropenia (6), alopecia (7), peripheral neuropathy (8) and cardiotoxicity (9), all of which significantly decrease the patient’s quality of life. Furthermore, older patients in the terminal stages may also be more intolerant to these adverse reactions.
Folium Artemisia argyi (FAA), commonly called wormwood, is a perennial herb belonging to Artemisia in the Asteraceae family and rich in volatile oils, polysaccharides, flavonoids and other trace elements. FAA has strong adaptability and distributes in most parts of China. It is also cultivated in Mongolia, Korea, Russia’s Far East and Japan. As a traditional Chinese herbal medicine, FAA has antipyretic, analgesic, and hemostatic effects (10). For thousands of years, it has been used internally to warm channels, arrest bleeding, dispel cold, and relieve pain, and is applied externally to eliminate dampness and relieve itching (11). Recently, owing to the various limitations of Western medicine, such as the toxicity and adverse side effects, increasing attention has been paid to the role of traditional Chinese medicine in the prevention and treatment of cancer (12). At the same time, Chinese herbs can target multiple points to achieve synergistic actions (13,14). According to pharmacology research, FAA contains multiple active chemical constituents, such as flavonoids, terpenoids, phenolic acids, and volatile oils (15,16), and exhibits a variety of effects, including anticancer, anti-inflammation, and anti-oxidation (17,18). For example, Shafi et al. suggested that FAA inhibited the proliferation and promoted apoptosis in breast cancer cells through Bcl-2 family proteins and the MEK/ERK pathway (10). It was also reported that FAA exhibited a dose-dependent inhibitory effect on hepatoma cells (11). However, although many studies have verified that FAA exerts remarkable antitumor functions, the underlying mechanisms have not yet been comprehensively understood (10,11,17,18).
It is widely known that herbal medicines include multi-component, multi-target, and multi-pathway features (19,20). Traditional Chinese medicine network pharmacology is a systematic research method based on the interaction network of herbs, compounds, targets, diseases, and genes (21). This approach emphasizes the integration of bioinformatics, systems biology, and pharmacology, which not only explains the complex interactions between herbs and diseases at a systematic level but also conforms to the systematic and holistic perspective of the traditional Chinese medicine theory (22,23). Thus, we utilized a network pharmacology approach in this study to explore the pharmacological mechanisms of FAA as a treatment for breast cancer. Firstly, we screened for active ingredients of FAA by estimating their oral bioavailability (OB) and drug-likeness (DL) (24). Next, we selected the common targets shared by the FAA compound targets and the breast cancer-related targets using two databases [GeneCards and Online Mendelian Inheritance in Man (OMIM)] and then constructed the network by investigating the potential interactions between the various target nodes. In addition, the protein-protein interaction (PPI) data were obtained from the Search Tool for the Retrieval of Interacting Genes (STRING) database, and enrichment analyses [Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)] were performed to explore the potential mechanisms of FAA against breast cancer. In summary, this study aimed to identify the potential targets and pathways of FAA as a treatment for breast cancer using the network pharmacology approach, and systematically elucidate the mechanisms of FAA in the treatment of breast cancer. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5769/rc).
Methods
Data preparation
Active ingredients and targets against breast cancer in FAA
FAA ingredients were acquired from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database, which serves as a systematic platform to study herbs, including the identification of compounds and the screening of compound targets (25). In addition, to identify the corresponding targets of FAA compounds against breast cancer, the TCMSP database was utilized to identify potential targets. Finally, nine active herbal ingredients of FAA were selected (Table 1) by linking the active ingredients of FAA to the breast cancer targets. A total of 236 targets of FAA compounds were obtained in total (the specific targets are not shown).
Table 1
| Mol ID | Mol name | 2D structure | OB (%) | DL |
|---|---|---|---|---|
| MOL001040 | (2R)-5,7-dihydroxy-2-(4-hydroxyphenyl) chroman-4-one |  |
42.36 | 0.21 |
| MOL001494 | Mandenol |  |
42 | 0.19 |
| MOL002883 | Ethyl oleate (NF) |  |
32.4 | 0.19 |
| MOL000358 | beta-sitosterol |  |
36.91 | 0.75 |
| MOL000449 | Stigmasterol |  |
43.83 | 0.76 |
| MOL005720 | 24-methylenecyloartanone |  |
41.11 | 0.79 |
| MOL005735 | dammaradienyl acetate |  |
44.83 | 0.83 |
| MOL005741 | cycloartenol acetate |  |
41.11 | 0.8 |
| MOL000098 | quercetin |  |
46.43 | 0.28 |
FAA, folium Artemisia argyi; Mol, molecular; OB, oral bioavailability; DL, drug-likeness.
Pharmacokinetic predictions
In a pharmaceutical study, ADME (absorption, distribution, metabolism, and excretion) is a critical pattern to identify (24). Therefore, we employed two major ADME-related properties, namely, OB and DL to search for potential bioactive compounds of FAA. Ingredients with OB ≥30% and DL ≥0.18 were considered to be suggested drug screening criteria. The screening criteria of OB ≥30% and DL ≥0.18 to select ingredients was set based on previous studies (26,27). This criterion allows for more accurate screening of active ingredients. Detailed information on all of the ingredients before screening is listed in Table S1.
Potential target genes of breast cancer
Breast cancer-related target genes data were gathered from the GeneCards and OMIM databases. The species was set to Homo sapiens. GeneCards is an extensive platform that provides insight into predicted and annotated human genes. All of the gene-centric data were collected from 150 web resources, including genetic, genomic, proteomic, transcriptomic, and functional information (28). Search strategy: we set the keyword as “breast cancer” and the score30 after logging in to Genecards. The detailed information is listed in Table S2.
The OMIM is a comprehensive, authoritative, and timely knowledgebase that links and catalogues all known diseases with a genetic component and provides further references to the genomic analyses of catalogued genes (29). Search strategy: we chose “gene map” on the website and then set the keyword as “breast cancer”. The detailed information is listed in Table S3.
PPI data
We acquired the PPI data from the STRING database, which defines PPI with confidence ranges for data scores (high >0.7; medium >0.4; low >0.15) (30). In this study, we selected a confidence score of >0.4 to construct our PPI network.
Network construction
The PPI network has been widely applied to display many different interactions between proteins in the context of complex diseases (23,31), including breast cancer, prostate cancer, lung cancer, gastric cancer, etc. In this study, we constructed the network as follows: (I) we acquired the targets shared by the FAA compound targets and the breast cancer-related targets; (II) we entered these targets into the STRING database and obtained the FAA against breast cancer targets PPI network; and (III) we exported the PPI results as a simple tabular text output (.tsv) and then imported the .tsv file into Cytoscape (version 3.6.1, National Resource for Network Biology (NRNB), USA) to reconstruct the network to achieve better visualization and understanding for further analysis (32).
GO and KEGG pathway enrichment analysis
In this study, we used the Cluster Profiler package of R3.5.2 (Bioconductor, China) to perform GO enrichment analysis of the targets; the higher the score, the greater the importance of the genes represented in the list (33). The Cluster Profiler package of R3.5.2 was also used to analyze the KEGG pathway enrichment of overlapping target genes. KEGG analysis was used to explore the biological pathways and potential biological functions based on the enrichment analysis of functional items (34). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Active ingredients
In this study, we acquired a total of nine active ingredients in FAA after ADME identification. Detailed information is shown in Table 1 (all Mol IDs (Bioconductor, China) could be tracked in the TCMSP database). All of the FAA compounds before screening are presented in Table S1.
FAA compound-target network
To further uncover the potential pharmacological mechanisms of FAA against breast cancer, target genes common to both the active ingredients of FAA and breast cancer were selected in different databases. A total of 75 genes (Figure S1) belonging to both the FAA target gene and breast cancer target gene networks were screened via Venn analysis (Figure 1A). The compound-target network is presented in Figure 1B and includes 82 nodes and 171 edges, with a network density of 0.051 and a network diameter of 3. Detailed information on this network is depicted in Table 2.
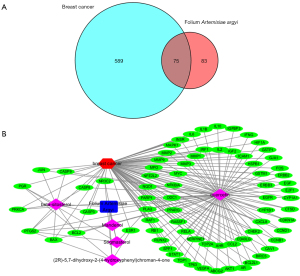
Table 2
| Network parameters | Values |
|---|---|
| Number of nodes | 82 |
| Network density | 0.051 |
| Network diameter | 3 |
| Network heterogeneity | 2.678 |
| Average number of neighbors | 4.146 |
| Characteristic path length | 2.035 |
| Shortest paths | 6,642 (100%) |
| Network centralization | 0.897 |
FAA, folium Artemisia argyi.
PPI network
To explore the underlying mechanisms of FAA as a therapy against breast cancer, a PPI network of the FAA compound targets against breast cancer was constructed by connecting the targets of the FAA compounds and breast cancer. First, we obtained a total of 75 target genes belonging to both the FAA target gene and breast cancer target gene and obtained target symbol names using UniProt. Next, all of these 75 target genes were imported into the STRING database to generate the PPI results (settings: Homo sapiens and confidence>0.4). The original STRING PPI network is presented in Figure S2. Next, we imported the PPI data generated in the STRING database into Cytoscape (version 3.6.1).
As shown in Figure 2A, this PPI network included 75 nodes and 1,247 edges, with a network diameter of 3, a clustering coefficient of 0.733, and an average number of 33.253 neighbors. The average node degree was 33.3 (the degree was indicated both by the different colors and the size of the circles). Detailed information on this network is displayed in Table 3. All target degrees were calculated using this network. In Figure 2B, the 10 targets with the greatest degrees were AKT1 (degree =67), MYC (degree =65), CASP3 (degree =63), EGFR (degree =62), JUN (degree =61), CCND1 (degree =60), VEGFA (degree =60), ESR1 (degree =59), MAPK1 (degree =57), and EGF (degree =55). As shown in Figure 2C, the cluster consisted of 68 nodes and 1,155 edges. The average node degree was 34 and the clustering coefficient was 0.77. AKT1 (The red diamond in Figure 2C) was the seed in this cluster and interacted with the other FAA targets.
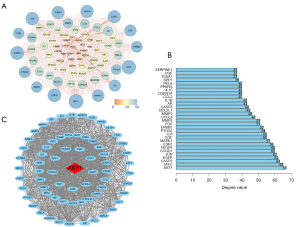
Table 3
| Network parameters | Values |
|---|---|
| Number of nodes | 75 |
| Number of edges | 1,247 |
| Network diameter | 3 |
| Clustering coefficient | 0.733 |
| Average number of neighbors | 33.253 |
| Average node degree | 33.3 |
FAA, folium Artemisia argyi.
GO enrichment
To further discuss the multiple mechanisms of FAA as a treatment against breast cancer, we conducted GO enrichment analysis on the 75 common targets shared by the FAA compound targets and the breast cancer-related targets (35). Specifically, the top 30 targets are as follows (Figure 2B): AKT1, MYC, CASP3, EGFR, JUN, CCND1, VEGFA, ESR1, MAPK1, EGF, IL6, PTGS2, ERBB2, FOS, MMP9, CXCL8, MMP2, BCL2L1, CASP8, AR, IL1B, CCL2, CDKN1A, IL10, PPARG, RELA, SPP1, ICAM1, PGR, and SERPINE1. The significantly enriched GO targets are presented (adjusted P value <0.001) in Figure 3. The top five GO enrichment targets included (I) transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding (GO:0000982); (II) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding (GO:0001228); (III) ubiquitin-like protein ligase binding (GO:0044389); (IV) cytokine receptor binding (GO:0005126); and (V) transcriptional activator activity, RNA polymerase II proximal promoter sequence-specific DNA binding (GO:0001077). Detailed GO enrichment information was shown in Table 4. Thus, we speculated that FAA probably executed its pharmacological effects on breast cancer by simultaneously involving these molecular functions.
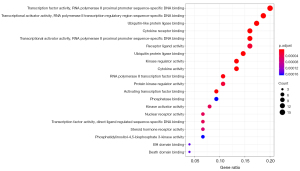
Table 4
| ID | Description | Count | Adjust P value |
|---|---|---|---|
| GO:0000982 | Transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 15 | <0.0001 |
| GO:0001228 | Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific DNA binding |
14 | <0.0001 |
| GO:0044389 | Ubiquitin-like protein ligase binding | 13 | <0.0001 |
| GO:0005126 | Cytokine receptor binding | 12 | <0.0001 |
| GO:0001077 | Transcriptional activator activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 12 | <0.0001 |
| GO:0048018 | Receptor ligand activity | 12 | <0.0001 |
| GO:0031625 | Ubiquitin protein ligase binding | 11 | <0.0001 |
| GO:0019207 | Kinase regulator activity | 10 | <0.0001 |
| GO:0005125 | Cytokine activity | 10 | <0.0001 |
| GO:0001085 | RNA polymerase II transcription factor binding | 8 | <0.0001 |
| GO:0019887 | Protein kinase regulator activity | 8 | <0.0001 |
| GO:0033613 | Activating transcription factor binding | 7 | <0.0001 |
| GO:0019902 | Phosphatase binding | 7 | 0.0002 |
| GO:0019209 | Kinase activator activity | 6 | <0.0001 |
| GO:0004879 | Nuclear receptor activity | 5 | <0.0001 |
| GO:0098531 | Transcription factor activity, direct ligand regulated sequence-specific DNA binding | 5 | <0.0001 |
| GO:0003707 | Steroid hormone receptor activity | 5 | <0.0001 |
| GO:0046934 | Phosphatidylinositol-4,5-bisphosphate 3-kinase activity | 5 | 0.0002 |
| GO:0051400 | BH domain binding | 3 | 0.0001 |
| GO:0070513 | Death domain binding | 3 | 0.0001 |
GO, Gene Ontology.
KEGG enrichment
We obtained a total of 74 pathways belonging to several categories, including human diseases, cellular processes, and drug resistance, among others. Among these, the top 30 significantly enriched KEGG targets are presented (adjusted P value <0.001) in Figure 4. In the cancer-related disease, prostate cancer (hsa05215), bladder cancer (hsa05219), pancreatic cancer (hsa05212), breast cancer (hsa05224), colorectal cancer (hsa05210), non-small cell lung cancer (hsa05223), small cell lung cancer (hsa05222), gastric cancer (hsa05226), endometrial cancer (hsa05226), renal cell carcinoma (hsa05211), thyroid cancer (hsa05216), and small cell lung cancer (hsa05222) data were processed using KEGG enrichment analysis. Detailed KEGG information is shown in Table 5.
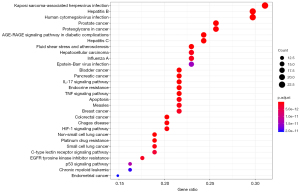
Table 5
| ID | Description | Count | Adjust P value |
|---|---|---|---|
| hsa05167 | Kaposi sarcoma-associated herpes virus infection | 23 | <0.0001 |
| hsa05161 | Hepatitis B | 22 | <0.0001 |
| hsa05163 | Human cytomegalovirus infection | 22 | <0.0001 |
| hsa04151 | PI3K-Akt signaling pathway | 21 | <0.0001 |
| hsa05215 | Prostate cancer | 19 | <0.0001 |
| hsa05205 | Proteoglycans in cancer | 19 | <0.0001 |
| hsa05206 | MicroRNAs in cancer | 19 | <0.0001 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 18 | <0.0001 |
| hsa05160 | Hepatitis C | 18 | <0.0001 |
| hsa04010 | MAPK signaling pathway | 18 | <0.0001 |
| hsa05165 | Human papillomavirus infection | 18 | <0.0001 |
| hsa05418 | Fluid shear stress and atherosclerosis | 17 | <0.0001 |
| hsa05225 | Hepatocellular carcinoma | 17 | <0.0001 |
| hsa05164 | Influenza A | 17 | <0.0001 |
| hsa05169 | Epstein-Barr virus infection | 17 | <0.0001 |
| hsa05166 | Human T-cell leukemia virus 1 infection | 17 | <0.0001 |
| hsa05219 | Bladder cancer | 16 | <0.0001 |
| hsa05212 | Pancreatic cancer | 16 | <0.0001 |
| hsa04657 | IL-17 signaling pathway | 16 | <0.0001 |
| hsa01522 | Endocrine resistance | 16 | <0.0001 |
| hsa04668 | TNF signaling pathway | 16 | <0.0001 |
| hsa04210 | Apoptosis | 16 | <0.0001 |
| hsa05162 | Measles | 16 | <0.0001 |
| hsa05224 | Breast cancer | 16 | <0.0001 |
| hsa05210 | Colorectal cancer | 15 | <0.0001 |
| hsa05142 | Chagas disease | 15 | <0.0001 |
| hsa04066 | HIF-1 signaling pathway | 15 | <0.0001 |
| hsa04218 | Cellular senescence | 15 | <0.0001 |
| hsa05152 | Tuberculosis | 15 | <0.0001 |
| hsa05170 | Human immunodeficiency virus 1 infection | 15 | <0.0001 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
As demonstrated in Figure 1B, quercetin was the most critical component of FAA, which was connected to the most targets. It is a flavonoid found in natural plants and exhibits a variety of activities such as antioxidant, anti-inflammatory, antiviral, and antimicrobial effects through multiple signal transduction pathways (35,36). Several studies have validated that quercetin can inhibit the progression of various tumors, including breast cancer (37), prostate cancer (38), gastric cancer (39), ovarian cancer (40), and colorectal cancer (41). Moreover, some studies have also reported that it not only has a synergistic effect when combined with chemotherapeutic or radiotherapy agents but can also mitigate the expected adverse side effects and toxic reactions (42,43).
In addition, our results showed that numerous targets were affected by two or more compounds. For instance, Prostaglandin G/H synthase 1 (PTGS1) and Prostaglandin G/H synthase 2 (PTGS2) were both modulated by quercetin, stigmasterol, mandenol, etc. Constitutive PTGS1 and inducible PTGS2 belong to two isozymes of PTGS, which have pivotal effects both as a peroxidase and a dioxygenase (44). Other studies have suggested that PTGS2 might inversely control the metastasis and chemoresistance of breast cancer via the regulation of EMT (Epithelial-mesenchymal transition), apoptosis, and senescence (45-47). Also, nuclear receptor coactivator 2 (NCOA2), a member of the p160 family, performs key roles in many different physiological and pathological processes, including cell growth, energy metabolism, endocrine regulation, and circadian rhythms (48). More importantly, NCOA2 gene expression plays crucial roles in the development, progression, and metastasis of malignant tumors, including breast cancer (49). In prostate cancer patients, the high expression of NCOA2 is more likely to relapse after androgen deprivation therapy (50).
Similarly, Beta-2 adrenergic receptor (ADRB2), Gamma-aminobutyric acid receptor subunit alpha-1 (GABRA1), Heat shock protein HSP 90, Progesterone receptor (PGR), and Sodium channel protein type 5 subunit alpha (SCN5A) could also be regulated by more than two active ingredients. In this study, we obtained an approximate observation of the relationship between these active ingredients and targets and also discovered the potential pharmacological effects of FAA from this network (Figure 1B).
Figure 2C intuitively indicated that AKT1 played an important role in connecting other nodes in this PPI network. It is well-known that the serine/threonine kinase, AKT1, one of the three isoforms of the Akt family, emerged as a downstream effector of PI3K (51). AKT inhibits apoptosis by suppressing the actions of BAD (BCL2 Associated Agonist of Cell Death) and caspase-9 (52). In breast cancer, AKT1 activation accelerates cell proliferation, while Akt1 inhibition promotes epithelial-to-mesenchymal transition (53).
Detailed GO enrichment information is shown in Table 4. We speculated that FAA probably executed its pharmacological effects on breast cancer by simultaneously involving these molecular functions. We further carried out KEGG (34) enrichment analysis on 75 common targets to clarify the integral regulation of FAA in the treatment of breast cancer. The results (shown in Table 5) indicated that FAA has a significant potential to treat a wide range of cancers, such as breast cancer (10), prostate cancer (54), bladder cancer (55), colorectal cancer (56), and gastric cancer (57), which is consistent with previous research. Furthermore, the results also verified that these signaling pathways remarkably enriched by potential targets of FAA in breast cancer were strongly associated with signal transduction, endocrine system, replication, repair, as well as cell growth and death, most of which played an essential role in the development and progression of cancers, such as the PI3K/AKT signaling pathway (hsa04151) (58), the MAPK signaling pathway (hsa04010) (59), the mammalian target of rapamycin (mTOR) signaling pathway (hsa04150) (60), apoptosis (hsa04210), and the cell cycle signaling pathway (hsa04110) (61). Therefore, we speculated that the underlying mechanism of FAA against breast cancer might be attributed to the coordinated regulation of several cancer-related pathways. About 30% of breast cancer patients who have poor prognosis show overexpression and amplification of HER2 gene. The high receptor concentration on the membranes of HER2 overexpressing cells can activate the PI3K/AKT and the RAS/RAF/MAPK pathways, which initiates cell proliferation, growth, survival, invasion and angiogenesis. It is considered that FAA is related to the above signaling pathways in breast cancer. FAA may inhibit the recurrence and metastasis of HER2-positive breast cancer patients, which needs further verification (62,63).
Although breast cancer patients get better after various treatments, most patients still have drug resistance and show disease progression. This process involves multiple signaling pathways. According to our results, FAA may be related to multiple signaling pathways in breast cancer. Quercetin was the most critical component of FAA, several studies have validated that quercetin can inhibit the progression of various tumors (38,41,42). Therefore, FAA may be considered as a therapeutic agent against various cancers including breast cancer in the future. The absence of experimental data is indeed the weakness of this study. In fact, the relevant experiments are carrying out to validate the representative active ingredients of FAA, but have not been completed. We are very pleased to report the experimental results in the subsequent studies.
Conclusions
At present, although many studies have verified that FAA exhibits arresting antitumor activities, the underlying mechanisms of its antitumor activities have not yet been fully elucidated. Network pharmacology emphasizes the integration of bioinformatics, systems biology, and pharmacology, which not only explain the complex interactions between diseases and Chinese herbs at a systematic level but also conform to the systematic and holistic perspective of traditional Chinese medicine theory (12). To better explore the pharmacological mechanisms of FAA as a treatment for breast cancer, we applied the network pharmacology approach to identify the potential mechanisms of FAA as a breast cancer treatment by compound-target network construction, PPI network, and GO and KEGG enrichment analyses. We took advantage of OB and DL to explore the potential active ingredients of FAA. At present, there are few studies on the pharmacokinetics of FAA. Choi et al. found that FAA exerted anticancer activities through the inhibition of cell growth and the induction of apoptosis in breast cancer cells (17).
In this study, we acquired nine active ingredients and 236 potential targets from FAA and validated a synergistic herb strategy featuring multi-component, multi-target, and multi-pathway characteristics. The compound-target network confirmed that quercetin served as the major ingredient in FAA. Moreover, the PPI network provided information concerning the source of the interactions. PPI analysis indicated that FAA had a significant effect on breast cancer by influencing the whole biological network, including targets such as AKT1, MYC, CASP3, EGFR, JUN, CCND1, VEGFA, ESR1, MAPK1, and EGF. The PPI cluster demonstrated that AKT1 was the seed, suggesting that AKT1 played a crucial role in connecting other nodes in the PPI network. Next, enrichment analysis indicated that FAA was strongly related to signal transduction, the endocrine system, replication and repair, and cell growth and death. The enrichment results also showed that the underlying mechanism of FAA against breast cancer might be attributed to the coordinated regulation of several cancer-related pathways, such as the MAPK and mTOR signaling pathways, among others.
In conclusion, this study applied a network approach demonstrating how FAA compounds alter different pathways against breast cancer, which was supplementary to other studies on drugs against breast cancer. Furthermore, we confirmed that FAA substantially influenced numerous breast cancer-related targets, a finding that was consistent with present cancer study trends showing that the occurrence and development of breast cancer is a result of the gradual accumulation of distinct genome modifications in cancer cells (64,65). We fully expect that our research can help to promote the employment of network pharmacology in uncovering the potential mechanisms of anticancer Chinese herbs and provide clues to assess the synergy of herbs in the treatment of other complex diseases, especially cancer.
Acknowledgments
Funding: This study was supported by Heilongjiang Science Foundation (Grant No. H2018046).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5769/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5769/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tao Z, Shi A, Lu C, et al. Breast Cancer: Epidemiology and Etiology. Cell Biochem Biophys 2015;72:333-8. [Crossref] [PubMed]
- Druesne-Pecollo N, Touvier M, Barrandon E, et al. Excess body weight and second primary cancer risk after breast cancer: a systematic review and meta-analysis of prospective studies. Breast Cancer Res Treat 2012;135:647-54. [Crossref] [PubMed]
- Hortobagyi GN, de la Garza Salazar J, Pritchard K, et al. The global breast cancer burden: variations in epidemiology and survival. Clin Breast Cancer 2005;6:391-401. [Crossref] [PubMed]
- Barzaman K, Karami J, Zarei Z, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol 2020;84:106535. [Crossref] [PubMed]
- Trayes KP, Cokenakes SEH. Breast Cancer Treatment. Am Fam Physician 2021;104:171-8.
- Nomura M, Morita Y, Kakiuchi A, et al. The association between chemotherapy-induced febrile neutropenia and breast cancer subtype in Japanese patients. Int J Clin Pharm 2020;42:7-10. [Crossref] [PubMed]
- Munzone E, Bagnardi V, Campennì G, et al. Preventing chemotherapy-induced alopecia: a prospective clinical trial on the efficacy and safety of a scalp-cooling system in early breast cancer patients treated with anthracyclines. Br J Cancer 2019;121:325-31. [Crossref] [PubMed]
- Avan R, Janbabaei G, Hendouei N, et al. The effect of pregabalin and duloxetine treatment on quality of life of breast cancer patients with taxane-induced sensory neuropathy: A randomized clinical trial. J Res Med Sci 2018;23:52. [Crossref] [PubMed]
- Cai F, Luis MAF, Lin X, et al. Anthracycline-induced cardiotoxicity in the chemotherapy treatment of breast cancer: Preventive strategies and treatment. Mol Clin Oncol 2019;11:15-23. [Crossref] [PubMed]
- Shafi G, Hasan TN, Syed NA, et al. Artemisia absinthium (AA): a novel potential complementary and alternative medicine for breast cancer. Mol Biol Rep 2012;39:7373-9. [Crossref] [PubMed]
- Liu R, Zhao J, He K, et al. Determination of Eupatilin in Folium artemisiae Argyi and Its Inhibitory Effect on Hepatoma Cells. Pharmacogn Mag 2018;14:129-33. [Crossref] [PubMed]
- Song Y, Wang H, Pan Y, et al. Investigating the Multi-Target Pharmacological Mechanism of Hedyotis diffusa Willd Acting on Prostate Cancer: A Network Pharmacology Approach. Biomolecules 2019;9:591. [Crossref] [PubMed]
- Zhu JY, Lavrik IN, Mahlknecht U, et al. The traditional Chinese herbal compound rocaglamide preferentially induces apoptosis in leukemia cells by modulation of mitogen-activated protein kinase activities. Int J Cancer 2007;121:1839-46. [Crossref] [PubMed]
- Garcia-Carbonero R, Supko JG. Current perspectives on the clinical experience, pharmacology, and continued development of the camptothecins. Clin Cancer Res 2002;8:641-61.
- Xia JX, Zhao BB, Zan JF, et al. Simultaneous determination of phenolic acids and flavonoids in Artemisiae Argyi Folium by HPLC-MS/MS and discovery of antioxidant ingredients based on relevance analysis. J Pharm Biomed Anal 2019;175:112734. [Crossref] [PubMed]
- Wang XJ, Deng YH, Zhang LP, et al. Identification and determination of phenolic acids and flavonoids in Artemisiae Argyi Folium by UPLC-DAD-MS. Zhongguo Zhong Yao Za Zhi 2019;44:983-9. [Crossref] [PubMed]
- Choi E, Kim G. Effect of artemisia species on cellular proliferation and apoptosis in human breast cancer cells via estrogen receptor-related pathway. J Tradit Chin Med 2013;33:658-63. [Crossref] [PubMed]
- Jeong MA, Lee KW, Yoon DY, et al. Jaceosidin, a pharmacologically active flavone derived from Artemisia argyi, inhibits phorbol-ester-induced upregulation of COX-2 and MMP-9 by blocking phosphorylation of ERK-1 and -2 in cultured human mammary epithelial cells. Ann N Y Acad Sci 2007;1095:458-66. [Crossref] [PubMed]
- Liu X, Wu J, Zhang D, et al. A Network Pharmacology Approach to Uncover the Multiple Mechanisms of Hedyotis diffusa Willd. on Colorectal Cancer. Evid Based Complement Alternat Med 2018;2018:6517034. [Crossref] [PubMed]
- Gao L, Hao J, Niu YY, et al. Network pharmacology dissection of multiscale mechanisms of herbal medicines in stage IV gastric adenocarcinoma treatment. Medicine (Baltimore) 2016;95:e4389. [Crossref] [PubMed]
- Lee D, Lee WY, Jung K, et al. The Inhibitory Effect of Cordycepin on the Proliferation of MCF-7 Breast Cancer Cells, and its Mechanism: An Investigation Using Network Pharmacology-Based Analysis. Biomolecules 2019;9:407. [Crossref] [PubMed]
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 2008;4:682-90. [Crossref] [PubMed]
- Li S, Fan TP, Jia W, et al. Network pharmacology in traditional chinese medicine. Evid Based Complement Alternat Med 2014;2014:138460. [Crossref] [PubMed]
- Barton HA, Pastoor TP, Baetcke K, et al. The acquisition and application of absorption, distribution, metabolism, and excretion (ADME) data in agricultural chemical safety assessments. Crit Rev Toxicol 2006;36:9-35. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Wan Y, Xu L, Liu Z, et al. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement Altern Med 2019;19:158. [Crossref] [PubMed]
- Xu T, Wang Q, Liu M. A Network Pharmacology Approach to Explore the Potential Mechanisms of Huangqin-Baishao Herb Pair in Treatment of Cancer. Med Sci Monit 2020;26:e923199. [Crossref] [PubMed]
- Rebhan M, Chalifa-Caspi V, Prilusky J, et al. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998;14:656-64. [Crossref] [PubMed]
- Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33:D514-7. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med 2013;11:110-20. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Jeong JH, An JY, Kwon YT, et al. Effects of low dose quercetin: cancer cell-specific inhibition of cell cycle progression. J Cell Biochem 2009;106:73-82. [Crossref] [PubMed]
- Yoshida M, Sakai T, Hosokawa N, et al. The effect of quercetin on cell cycle progression and growth of human gastric cancer cells. FEBS Lett 1990;260:10-3. [Crossref] [PubMed]
- Li X, Zhou N, Wang J, et al. Quercetin suppresses breast cancer stem cells (CD44(+)/CD24(-)) by inhibiting the PI3K/Akt/mTOR-signaling pathway. Life Sci 2018;196:56-62. [Crossref] [PubMed]
- Ward AB, Mir H, Kapur N, et al. Quercetin inhibits prostate cancer by attenuating cell survival and inhibiting anti-apoptotic pathways. World J Surg Oncol 2018;16:108. [Crossref] [PubMed]
- Shang HS, Lu HF, Lee CH, et al. Quercetin induced cell apoptosis and altered gene expression in AGS human gastric cancer cells. Environ Toxicol 2018;33:1168-81. [Crossref] [PubMed]
- Shafabakhsh R, Asemi Z. Quercetin: a natural compound for ovarian cancer treatment. J Ovarian Res 2019;12:55. [Crossref] [PubMed]
- Darband SG, Kaviani M, Yousefi B, et al. Quercetin: A functional dietary flavonoid with potential chemo-preventive properties in colorectal cancer. J Cell Physiol 2018;233:6544-60. [Crossref] [PubMed]
- Brito AF, Ribeiro M, Abrantes AM, et al. Quercetin in Cancer Treatment, Alone or in Combination with Conventional Therapeutics? Curr Med Chem 2015;22:3025-39. [Crossref] [PubMed]
- Daker M, Ahmad M, Khoo AS. Quercetin-induced inhibition and synergistic activity with cisplatin - a chemotherapeutic strategy for nasopharyngeal carcinoma cells. Cancer Cell Int 2012;12:34. [Crossref] [PubMed]
- Devi GR. siRNA-based approaches in cancer therapy. Cancer Gene Ther 2006;13:819-29. [Crossref] [PubMed]
- Xu H, Lin F, Wang Z, et al. CXCR2 promotes breast cancer metastasis and chemoresistance via suppression of AKT1 and activation of COX2. Cancer Lett 2018;412:69-80. [Crossref] [PubMed]
- Gan L, Qiu Z, Huang J, et al. Cyclooxygenase-2 in tumor-associated macrophages promotes metastatic potential of breast cancer cells through Akt pathway. Int J Biol Sci 2016;12:1533-43. [Crossref] [PubMed]
- Sharma B, Nawandar DM, Nannuru KC, et al. Targeting CXCR2 enhances chemotherapeutic response, inhibits mammary tumor growth, angiogenesis, and lung metastasis. Mol Cancer Ther 2013;12:799-808. [Crossref] [PubMed]
- Lin Z, Yang F, Lu D, et al. Knockdown of NCOA2 Inhibits the Growth and Progression of Gastric Cancer by Affecting the Wnt Signaling Pathway-Related Protein Expression. Technol Cancer Res Treat 2020;19:1533033820928072. [Crossref] [PubMed]
- Cai M, Liang X, Sun X, et al. Nuclear Receptor Coactivator 2 Promotes Human Breast Cancer Cell Growth by Positively Regulating the MAPK/ERK Pathway. Front Oncol 2019;9:164. [Crossref] [PubMed]
- Qin J, Lee HJ, Wu SP, et al. Androgen deprivation-induced NCoA2 promotes metastatic and castration-resistant prostate cancer. J Clin Invest 2014;124:5013-26. [Crossref] [PubMed]
- Liu X, Wu J, Zhang D, et al. Network Pharmacology-Based Approach to Investigate the Mechanisms of Hedyotis diffusa Willd. in the Treatment of Gastric Cancer. Evid Based Complement Alternat Med 2018;2018:7802639. [Crossref] [PubMed]
- Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999;96:857-68. [Crossref] [PubMed]
- Li W, Hou JZ, Niu J, Xi ZQ, Ma C, Sun H, et al. Akt1 inhibition promotes breast cancer metastasis through EGFR-mediated beta-catenin nuclear accumulation. Cell Commun Signal 2018;16:82.
- Michaelsen FW, Saeed ME, Schwarzkopf J, et al. Activity of Artemisia annua and artemisinin derivatives, in prostate carcinoma. Phytomedicine 2015;22:1223-31. [Crossref] [PubMed]
- Khlifi D, Sghaier RM, Amouri S, et al. Composition and anti-oxidant, anti-cancer and anti-inflammatory activities of Artemisia herba-alba, Ruta chalpensis L. and Peganum harmala L. Food Chem Toxicol 2013;55:202-8. [Crossref] [PubMed]
- Lian G, Li F, Yin Y, et al. Herbal extract of Artemisia vulgaris (mugwort) induces antitumor effects in HCT-15 human colon cancer cells via autophagy induction, cell migration suppression and loss of mitochondrial membrane potential. J BUON 2018;23:73-8.
- Mousavi B, Tafvizi F, Zaker Bostanabad S. Green synthesis of silver nanoparticles using Artemisia turcomanica leaf extract and the study of anti-cancer effect and apoptosis induction on gastric cancer cell line (AGS). Artif Cells Nanomed Biotechnol 2018;46:499-510. [Crossref] [PubMed]
- Afify SM, Oo AKK, Hassan G, et al. How can we turn the PI3K/AKT/mTOR pathway down? Insights into inhibition and treatment of cancer. Expert Rev Anticancer Ther 2021;21:605-19. [Crossref] [PubMed]
- Rezatabar S, Karimian A, Rameshknia V, et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J Cell Physiol 2019; Epub ahead of print. [Crossref]
- Sadeghalvad M, Mansouri K, Mohammadi-Motlagh HR, et al. Long non-coding RNA HOTAIR induces the PI3K/AKT/mTOR signaling pathway in breast cancer cells. Rev Assoc Med Bras (1992) 2022;68:456-62. [Crossref] [PubMed]
- Heydarnezhad Asl M, Pasban Khelejani F, Bahojb Mahdavi SZ, et al. The various regulatory functions of long noncoding RNAs in apoptosis, cell cycle, and cellular senescence. J Cell Biochem 2022;123:995-1024. [Crossref] [PubMed]
- Lee-Hoeflich ST, Crocker L, Yao E, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res 2008;68:5878-87. [Crossref] [PubMed]
- Dittrich A, Gautrey H, Browell D, et al. The HER2 Signaling Network in Breast Cancer--Like a Spider in its Web. J Mammary Gland Biol Neoplasia 2014;19:253-70. [Crossref] [PubMed]
- Chang HT, Li SC, Ho MR, et al. Comprehensive analysis of microRNAs in breast cancer. BMC Genomics 2012;13:S18. [Crossref] [PubMed]
- Peng X, Chang H, Gu Y, et al. 3,6-Dihydroxyflavone Suppresses Breast Carcinogenesis by Epigenetically Regulating miR-34a and miR-21. Cancer Prev Res (Phila) 2015;8:509-17. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


