
Ferroptosis-related genes with post-transcriptional regulation mechanisms in hepatocellular carcinoma determined by bioinformatics and experimental validation
Highlight box
Key findings
• Our findings identified new and yet-unrecognized regulatory molecular mechanisms of ferroptosis in hepatocellular carcinoma.
What is known and what is new?
• Limited evidence suggests that the modifications of m1A/m5C/m6A participate in regulating ferroptotic cell death of hepatocellular carcinoma. Nonetheless, mRNA modifications of most ferroptosis-related genes remain indistinct in hepatocellular carcinoma.
• The present study suggests that the ferroptosis-related genes preliminary elucidate the molecular mechanisms of HCC.
What is the implication, and what should change now?
• The research contributes to a better understanding of molecular mechanisms of ferroptosis in hepatocellular carcinoma, so we should shed light on the post-transcriptional regulation in hepatocellular carcinoma.
Introduction
Liver cancer is the third leading cause of cancer death worldwide in 2021 (1). Hepatocellular carcinoma (HCC) is the most common primary liver cancer (2). Despite the expanded implementation of resection and liver transplant globally, approximately 50–60% of HCC patients will eventually receive palliative therapy (3). Systemic molecular therapy has become the mainstay treatment for advanced-stage patients. It has been exciting time in the systemic therapy field for HCC. Sorafenib as single agent available for decades has limited survival benefits (4). Additionally, immunotherapy is revolutionizing the management of HCC (5). Despite this, only a minority of patients benefit from immunotherapy (6), and novel therapeutic strategies are therefore urgently required.
Successful circumvention of cell death regulation is the hallmark of tumorigenesis, leading to unlimited replication and immortality of cells. Ferroptosis is a form of iron-dependent cell death with increased free iron and massive lipid peroxidation. It is morphologically, genetically, and biochemically different from other types of cell death (apoptosis, necroptosis, pyroptosis, etc.) (7). The discovery of ferroptosis offers insights into cancer research. Dysregulated metabolic pathways and impaired iron homeostasis participate in mediating the progression of HCC through ferroptosis (8). Accumulated evidence demonstrates that ferroptosis is post-transcriptionally regulated in cancer cells (9). Circular RNAs (circRNAs) are a class of non-coding RNA molecules with tissue- or cancer-specific expression patterns (10). Most exert crucial functions in cancer development and progression via varying mechanisms of action, and have potential clinical implication and utility (11). Several circRNAs have been experimentally verified to participate in the ferroptosis of HCC cells. For instance, circ0097009 serves as a competing endogenous RNA to mediate the expression of SLC7A11, which is a critical regulator of ferroptosis, through sponging miR-1261 in HCC (9). CircIL4R attenuates ferroptosis in HCC via modulating miR-541-3p/glutathione peroxidase 4 (GPX4) signaling (12). Modifications of mRNAs mainly include N1-methyladenosine (m1A), 5-methylcytosine (m5C), and N6-methyladenosine (m6A), which form the epitranscriptome (13). The modification of m1A affects the first nitrogen atom of the adenine base and is positively charged under physiological conditions, which is deposited via “writers” (TRMT6/10C/61A/61B), erased via “erasers” (ALKBH1/3), and recognized via “readers” (YTHDF1/2/3 and YTHDC1) (14). The modification m5C has long been analyzed as an epigenetic modification in DNA. Recently, m5C was found to exist in mRNA of eukaryotic cells, which is deposited via “writers” (DNMT1/3A/3B), erased via “erasers” (TET1/2/3), and recognized via “readers” (MBD1/2/3/4, MeCP2, NEIL1, NTHL1, SMUG1, TDG, UHRF1/2, UNG, ZBTB4/33/38, etc.) (15). The modification m6A, which involves methylation at the sixth nitrogen atom of RNA base A, is the most abundant internal mRNA modification in eukaryotic cells. It is deposited via the m6A methyltransferase complex (comprising METTL3/14/16, WTAP, KIAA1429, and RBM15/15B), erased via demethylases (FTO and ALKBH5), and recognized via binding proteins (such as YTHDF1/2/3, YTHDC1/2, and IGF2BP1/2/3) (16). Limited evidence suggests that the modifications of m1A/m5C/m6A participate in regulating ferroptotic cell death of HCC. For instance, hypoxia attenuates ferroptosis of HCC by suppressing METTL14-triggered YTHDF2-dependent loss of SLC7A11 activity (17). Nonetheless, mRNA modifications of most ferroptosis-related genes remain indistinct in HCC.
In the present study, we first screened ferroptosis-related genes and their post-transcriptional regulation mechanisms, mainly comprising circRNA/miRNA and m1A/m5C/m6A RNA modifications in HCC. Further analysis demonstrated their mechanisms in HCC. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5750/rc).
Methods
Data acquisition
The circRNA microarrays (normalized series matrix files) of 3 paired HCC patient plasma and healthy control samples were acquired from the Gene Expression Omnibus (GEO) repository with accession number GSE166678 for the dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166678) on the GPL28148 platform. The mRNA sequencing data (counts) and microRNA (miRNA) normalized sequencing data [reads per million miRNAs mapped (RPM)] of 374 HCC and 50 normal specimens were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data preprocessing
Because circRNA microarray data were log2 normalized, no data preprocessing was performed. The mRNA sequencing data (counts) were firstly converted into counts per million (CPM) values using the edgeR package (18), and then the mRNAs whose CPM sample mean <10 in the matrix were eliminated to obtain the final expression matrix for subsequent analysis. Normalized RPM sequencing data for miRNAs were log2 normalized.
Differential expression analysis
Differential expression of circRNAs or miRNAs between HCC and control specimens was calculated using the limma package (19). Meanwhile, differential expression analysis of mRNAs between HCC and control specimens was implemented with the edgeR package (18). The threshold was set to |fold change| >1.5 and adjusted P<0.05.
Functional enrichment analysis
By utilizing enrichGO and enrichKEGG functions of the clusterProfiler package (20), functional enrichment analysis for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of differentially expressed mRNAs was implemented on the basis of hypergeometric distribution. To prevent a high false discovery rate (FDR) in multiple tests, the q-value was also computed for FDR control.
Gene set enrichment analysis (GSEA) is a reliable approach for inferring biological functions of a specific gene set through calculating the overlapped previously defined gene sets (21). In accordance with the median expression value of each ferroptosis-related gene, HCC samples were classified into high- and low-expression groups. The differences in GO biological process terms and KEGG pathways were compared between groups.
Development of a ferroptosis score and screening ferroptosis-related genes
The ferroptosis gene set (comprising FANCD2, NCOA4, TFRC, PHKG2, HSBP1, ACO1, FTH1, STEAP3, NFS1, IREB2, HMOX1, MT1G, ACSL4, AKR1C1, AKR1C2, AKR1C3, ALOX15, ALOX5, ALOX12, CARS1, CBS, CISD1, CS, DPP4, GPX4, HMGCR, LPCAT3, FDFT1, ACSL3, PEBP1, ZEB1, SQLE, FADS2, ACSF2, PTGS2, ACACA, GCLC, SLC7A11, KEAP1, NQO1, ABCC1, CHAC1, GSS, GCLM, NFE2L2, GLS2, SLC1A5, GOT1, G6PD, PGD, ATP5MC3, CD44, HSPB1, CRYAB, RPL8, SAT1, TP53, EMC2, AIFM2, and NOX1) was acquired from previously published literature. The ferroptosis score was computed utilizing the gene set variation analysis (GSVA) package on the basis of expression profiling of the ferroptosis gene set (22). Through the psych package, correlation analysis of the ferroptosis score with differentially expressed mRNAs was performed. In accordance with the correlation coefficient, the top 10 genes were selected as ferroptosis-related genes.
Prediction of circRNA-miRNA and miRNA-mRNA relationships
The circRNA and miRNA pairs were predicted using the Starbase database (http://starbase.sysu.edu.cn/) (23). Meanwhile, miRNA-mRNA relationships were predicted through the miRMap (http://cegg.unige.ch/mirmap) (24), miRanda (http://www.microrna.org) (25), miRDB (http://mirdb.org) (26), TargetScan (targetscan.org) (27), and miTarBase (http://miRTarBase.cuhk.edu.cn/) (28) databases. An miRNA-mRNA relationship pair in at least 4 databases was finally determined. The circRNA-miRNA-mRNA networks were established by applying the Cytoscape software (29).
Associations of ferroptosis-related genes and m1A/m5C/m6A regulators
The regulators of m1A/m5C/m6A RNA modifications were acquired from previously published literature (30). Pearson correlation was carried out to estimate the associations of ferroptosis-related genes and m1A/m5C/m6A regulators across HCC.
Immune cell infiltration analysis
The CIBERSORT approach (http://cibersort.stanford.edu/) (31) was applied to characterize cell compositions of HCC and normal tissues on the basis of mRNA expression profiling. The leukocyte gene signature matrix LM22 was utilized as a reference set, comprising 547 genes that distinguished 22 human hematopoietic cell populations (7 T cell types, naïve and memory B cells, plasma cells, NK cells, and myeloid subsets). Samples with CIBERSORT P<0.05 were used for subsequent analysis.
Cell culture and transfections
L-02, SMMC7721, and HepG2 (China Center Type Culture Collection) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37 ℃ with 5% CO2. To silence NUDCD1 expression, SMMC7721 and HepG2 cells were transfected with 100 nM small interfering RNA (siRNA) of NUDCD1 or scramble siRNA for 48 h using Lipofectamine 2000 (Sigma-Aldrich, USA).
RT-qPCR
Total RNA from L-02, SMMC7721, or HepG2 cells was extracted with Trizol reagent (Solarbio, China). Subsequently, RNA was converted to cDNA, followed by RT-PCR (Takara, Japan). The primer sequences included: NUDCD1, 5'-AAAACCACGAGAGGTGTTTCG-3' (forward), 5'-CTGACAAGGTAACCCAGGTAGA-3' (reverse); GAPDH, 5'-CTGGGCTACACTGAGCACC-3' (forward), 5'-AAGTGGTCGTTGAGGGCAATG-3' (reverse). The relative expression was computed via the 2-ΔΔCt approach with GAPDH as an internal control.
Western blot
Total protein from L-02, SMMC7721, or HepG2 cells was extracted using a protein extraction kit (Solarbio, China). The protein concentration was quantified with the Bradford approach. Protein samples (50 µg) were separated via denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transference onto polyvinylidene fluoride membranes. Membranes were blocked by 5% skim milk then incubated with primary antibodies targeting NUDCD1 (1/5,000; ab126902; Abcam, USA), GAPDH (1/5,000; ab199553), GPX4 (1/2,000; ab252833), and ferritin heavy chain 1 (FTH1) (1/2,000; ab75972), followed by a secondary antibody. Protein bands were developed with an enhanced chemiluminescence reagent.
Cell viability assay
Cell viability was assayed with Cell Counting Kit-8 (CCK-8) (Dojindo, Japan). SMMC7721 or HepG2 cells were seeded onto a 96-well plate and maintained for 0, 24, 48, 72, and 96 h. Subsequently, 10 µL CCK-8 solution was added to each well. The absorbance values at 450 nm were quantified with a microplate reader.
Glutathione persulfide (GSSG)/glutathione (GSH) assays
GSSG and total GSH levels were measured in the supernatants of SMMC7721 or HepG2 cells in accordance with the manufacturer’s protocol of corresponding kits (Nanjing Jiancheng Bioengineering Institute, China).
Statistical analysis
Data are displayed as mean ± standard deviation. Differences between groups were compared with Student’s t-test, the Wilcoxon test, or one-way analysis of variance (ANOVA). Overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS), or progression-free survival (PFS) were determined via the survival package. Correlations between variables were evaluated with the Pearson correlation test. All statistical analyses were implemented with R (version 3.6.3).
Results
Screening differentially expressed circRNAs, miRNAs, and mRNAs between HCC and control specimens
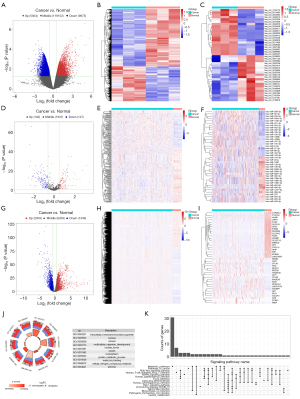
We acquired circRNA microarrays of 3 paired HCC patient plasma and healthy control samples from the GSE166678 dataset. According to the threshold of |fold change| >1.5 and adjusted P<0.05, 5,924 up-regulated circRNAs and 9,675 down-regulated circRNAs were found in HCC compared to control specimens (Figure 1A,1B). Figure 1C lists the top 20 up- and down-regulated circRNAs. We also acquired the mRNA and miRNA sequencing data of 374 HCC and 50 normal specimens from TCGA database. With the same threshold, we determined 102 up-regulated and 127 down-regulated miRNAs in HCC compared with controls (Figure 1D,1E). The top 20 up- and down-regulated miRNAs are separately displayed in Figure 1F. Additionally, 2,003 mRNAs were up-regulated and 1,344 were down-regulated in HCC compared with controls (Figure 1G,1H). A heatmap was used to visualize the top up- and down-regulated mRNAs (Figure 1I). Differentially expressed mRNAs participated in metabolic processes (Figure 1J) as well as tumorigenic pathways (Figure 1K; pathways in cancer).
Development of the ferroptosis score and screening ferroptosis-related genes
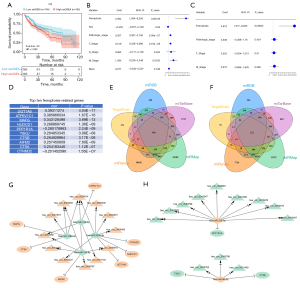
Using the GSVA approach, we computed the ferroptosis score for HCC on the basis of the ferroptosis gene set. With the median value of the ferroptosis score, we classified HCC patients into high- and low-ferroptosis score groups. Subsequently, we evaluated the survival difference between groups. Compared with the low-ferroptosis score group, a worse OS outcome was observed in the high-ferroptosis score group (Figure 2A). Univariate and multivariate cox regression analyses were conducted to investigate the associations of ferroptosis score and clinicopathological parameters with the OS of HCC cases. As illustrated in Figure 2B, ferroptosis score, pathological stage, T stage, and M stage were significantly linked to poorer OS. Further analysis demonstrated the independency of ferroptosis score in the OS of HCC cases (Figure 2C). Correlation analysis of ferroptosis score with differentially expressed mRNAs was conducted. In accordance with the correlation coefficient, the top 10 ferroptosis-related genes were determined, comprising UGT1A6, ATP6V1C1, MAFG, NUDCD1, PPP1R1A, TSKU, CTSB, AIFM2, CTSA, and CTNND2 (Figure 2D).
Ferroptosis-related genes regulated by circRNA-miRNA networks
Through the miRMap, miRanda, miRDB, TargetScan, and miTarBase databases, we predicted miRNA-mRNA relationships. Relationship pairs that appeared in at least 4 databases were finally determined. As a result, we obtained 944 pairs of down-regulated miRNAs and up-regulated mRNAs (Figure 2E) as well as 974 pairs of up-regulated miRNAs and down-regulated mRNAs (Figure 2F). Meanwhile, we acquired 3493 pairs of up-regulated miRNAs and down-regulated circRNAs as well as 2847 pairs of down-regulated miRNAs and up-regulated circRNAs. Through integrating circRNA-miRNA and miRNA-ferroptosis-related mRNA pairs, we established 2 networks (up-regulated circRNA/down-regulated miRNA/up-regulated ferroptosis-related mRNA, Figure 2G; and down-regulated circRNA/up-regulated miRNA/down-regulated ferroptosis-related mRNA, Figure 2H). Altogether, the above data unveiled ferroptosis-related genes regulated by circRNA-miRNA networks.
Ferroptosis-related genes regulated by m1A/m5C/m6A modifications
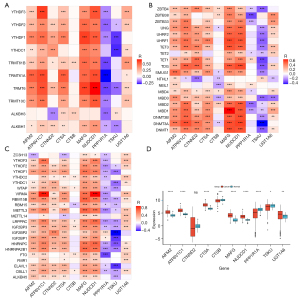
We further investigated the post-transcriptional regulatory mechanisms of ferroptosis-related genes by mRNA modifications. UGT1A6, ATP6V1C1, MAFG, NUDCD1, AIFM2, CTSA, and CTNND2 exhibited positive associations with most m1A, m5C, and m6A regulators (Figure 3A-3C). Meanwhile, PPP1R1A and TSKU were negatively correlated with most m1A, m5C, and m6A regulators. CTSB was positively or negatively correlated with several m1A, m5C, and m6A regulators. Altogether, these ferroptosis-related genes were post-transcriptionally regulated by m1A/m5C/m6A modifications. Their mRNA expression was compared between HCC and normal samples. AIFM2, ATP6V1C1, CTSA, MAFG, NUDCD1, and UGT1A6 displayed up-regulated expression in HCC compared with normal samples (Figure 3D). Meanwhile, CTSB, PPP1R1A, and TSKU expression was down-regulated in HCC compared with controls. No significant difference in CTNND2 was observed between HCC and normal samples.
Associations of ferroptosis-related genes with immune cell compositions within the tumor microenvironment
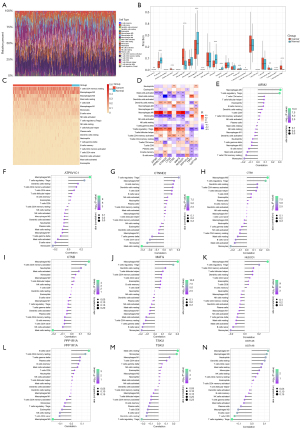
Through employing the CIBERSORT algorithm, 22 immune cell compositions were inferred in HCC and normal tissues (Figure 4A). There were marked differences in most immune cell compositions between HCC and normal tissues (Figure 4B,4C). Specifically, HCC tissues exhibited higher infiltration levels of resting dendritic cells, M0 macrophages, follicular helper T cells, and regulatory T cells (Tregs) than normal tissues. Meanwhile, lower infiltration levels of naïve B cells, M2 macrophages, monocytes, neutrophils, and plasma cells were observed in HCC compared with normal tissues. Correlation analysis of ferroptosis-related genes with immune cell compositions was further evaluated across HCC tissues. The results demonstrated that ferroptosis-related genes exhibited significant associations with most immune cells, especially macrophages M0 and Tregs (Figure 4C-4N).
Biological processes and pathways involved in ferroptosis-related genes
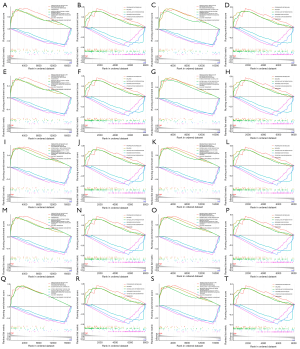
Biological processes and pathways involved in ferroptosis-related genes (UGT1A6, ATP6V1C1, MAFG, NUDCD1, PPP1R1A, TSKU, CTSB, AIFM2, CTSA, and CTNND2) were analyzed with GSEA. The results demonstrated that most ferroptosis-related genes were positively linked to the production of molecular mediators involved in inflammatory responses, innervation, negative regulation of bone mineralization, propanoate metabolism, malaria, and osteoclast differentiation, and were negatively associated with the interferon-mediated signaling pathway, sulfate transport, intra-S DNA damage checkpoint, arginine biosynthesis, other glycan degradation, and ribosome (Figure 5A-5T).
Prognostic implication of ferroptosis-related genes in HCC
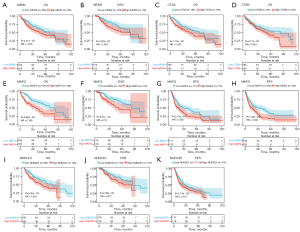
Among ferroptosis-related genes, up-regulated AIFM2 expression was positively correlated with the OS and DSS of HCC cases (Figure 6A,6B). High CTSA or CTSB expression contributed to poor OS outcomes (Figure 6C,6D). Patients with down-regulated MAFG expression had significant advantages in OS, DSS, DFS, and PFS outcomes (Figure 6E-6H). Additionally, lower OS, DSS, and DFS outcomes were observed in patients with high NUDCD1 expression (Figure 6I-6K). The above evidence demonstrated the prognostic implication of ferroptosis-related genes in HCC.
NUDCD1 suppression attenuates the proliferative capacity of HCC cells
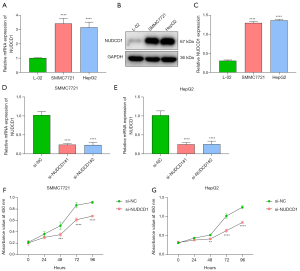
NUDCD1 is an oncogene commonly up-regulated in several human cancer types, but its biological implication in HCC is still unknown. The expression of the ferroptosis-related gene NUDCD1 was verified in L-02, SMMC7721, and HepG2 cells. In comparison to L-02 cells, NUDCD1 exhibited higher expression in SMMC7721 and HepG2 cells (Figure 7A-7C). Subsequently, si-NUDCD1 was transfected into SMMC7721 and HepG2 cells, and the results confirmed that NUDCD1 was knocked out (Figure 7D,7E). By utilizing CCK-8, a cell viability assay was performed. As a result, NUDCD1 suppression remarkably attenuated the proliferative capacity of HCC cells (Figure 7F,7G).
NUDCD1 suppression facilitates the ferroptosis of HCC cells
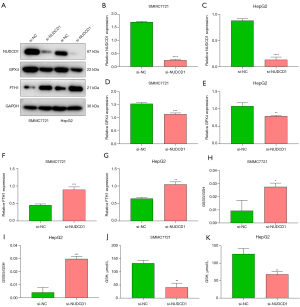
Western blots also demonstrated the knockdown of NUDCD1 in both SMMC7721 and HepG2 cells following si-NUDCD1 transfection (Figure 8A-8C). GPX4 utilizes GSH to protect cells from ferroptosis via eliminating phospholipid peroxides (32). In NUDCD1-knockout SMMC7721 and HepG2 cells, GPX4 expression was markedly attenuated (Figure 8D,8E). FTH1 is a crucial determinant for ferroptosis. Higher FTH1 expression was observed in NUDCD1-knockout SMMC7721 and HepG2 cells (Figure 8F,8G). Additionally, in both SMMC7721 and HepG2 cells, NUDCD1 suppression markedly elevated GSSG/GSH and mitigated GSH (Figure 8H-8K). Altogether, NUDCD1 suppression facilitated the ferroptosis of HCC cells.
Discussion
HCC represents a global health issue, with rising incidence and mortality. Ferroptosis is a newly discovered form of iron-dependent cell death, characterized by iron-dependent lipid peroxidation, loss of the endogenous antioxidant GSH, and altered mitochondrial morphology (33). Here, we computed a ferroptosis score on the basis of expression profiling of the ferroptosis gene set. Consistent with previous research (34), the ferroptosis score correlated with the poor OS of HCC cases, which was independent of other clinicopathological parameters. The present study determined 10 ferroptosis-related genes, namely UGT1A6, ATP6V1C1, MAFG, NUDCD1, PPP1R1A, TSKU, CTSB, AIFM2, CTSA, and CTNND2. Among them, ATP6V1C1 has been identified as an iron metabolism-related and methylated gene in HCC (35). MAFG possesses prognostic implications and correlates with ferroptosis in bladder cancer (36). CTSB acts as a promoter of ferroptosis (37). AIFM2 is a GSH-independent ferroptotic suppressor (38). Many studies have shown that alterations of m6A modification affect the development and progression of HCC, and they have important implications in the diagnosis, treatment and prognosis of HCC (39,40). Preliminary evidence suggests that m6A/m1A/m5C regulated genes play important biological roles in the progression of HCC, which was associated with poor prognosis and survival of HCC patients (41). Further analysis demonstrated their post-transcriptional regulation mechanisms (circRNA/miRNA and m1A/m5C/m6A RNA modifications) in HCC.
Iron-dependent cell death of tumor cells can cause the release of intracellular damage-associated molecular pattern (DAMP), which plays a dual role in anti-tumor immunity (42). On the one hand, DAMP brings out immunogenic cell death and enhances the anti-tumor function of dendritic cells and CD8+T cells, while CD8+T cells can in turn secrete IFN- γ to promote the occurrence of iron-dependent cell death (43,44); On the other hand, tumor cells release special DAMP, which stimulate TAM to transform into M2 type anti-inflammatory and tumor-promoting phenotype, thus promoting tumor occurrence and progression (45).
The composition of the immune microenvironment is the result of the interaction of immune suppressor cells, immune effector cells, cytokine milieu, and tumor cell intrinsic signaling pathways (2). Ferroptosis-related genes were significantly linked to most immune cells, especially M0 macrophages and Tregs. Most ferroptosis-related genes exhibited positive associations with the production of molecular mediators involved in inflammatory responses, innervation, negative regulation of bone mineralization, propanoate metabolism, malaria, and osteoclast differentiation, and displayed negative correlations with the interferon-mediated signaling pathway, sulfate transport, intra-S DNA damage checkpoint, arginine biosynthesis, other glycan degradation, and ribosome, indicative of the crucial functions of these ferroptosis-related genes in diverse biological processes and pathways. Among the 10 ferroptosis-related genes, up-regulated AIFM2 expression contributed to undesirable OS and DSS; high CTSA and CTSB expression correlated with poor OS outcome; down-regulated MAFG expression was positively associated with unfavorable OS, DSS, DFS, and PFS outcomes; and high NUDCD1 expression was linked to worse OS, DSS, and DFS outcomes, demonstrating the crucial prognostic implications of the above ferroptosis-related genes in HCC.
NUDCD1 is linked to Barcelona Clinic Liver Cancer (BCLC) staging and OS of HCC cases (46). Consistently, the present evidence showed the up-regulation of NUDCD1 expression in HCC tissues as well as the positive associations of NUDCD1 expression with poor OS, DSS, and DFS outcomes for HCC patients. NUDCD1 up-regulation was also observed in HCC cells compared with normal hepatocytes. Depletion of NUDCD1 activity attenuated the viability of HCC cells, indicative of the crucial function of NUDCD1 in HCC progression. Previous research reported the biological implications of NUDCD1 in other cancer types. For instance, loss of NUDCD1 mitigates the proliferation and metastases of pancreatic cancer (47) and colorectal cancer (48) through the epithelial-mesenchymal transition (EMT) process for non-small cell lung cancer cells through activating IGF1R-ERK1/2 (49). GPX4, a member of the GSH peroxidase family, reduces lipid peroxidase in cells, thereby aiding in cell survival (50). Depletion of GPX4 activity enhances phospholipid hydroperoxide and facilitates lipoxygenase-mediated lipid peroxidation, eventually resulting in ferroptotic cell death (51). GPX4 expression is markedly up-regulated in HCC tissues compared with controls (52), and is linked to HCC (53). Sorafenib, a multiple oncogenic kinase inhibitor, induces ferroptosis in HCC. Nevertheless, evidence suggests that several HCC cell lines exhibit lower sensitivity to sorafenib-induced ferroptosis. Suppression of GPX4 may sensitize HCC cells to sorafenib-induced ferroptosis (54). FTH1 is crucial for iron metabolism and ferroptosis and exhibits up-regulation in HCC cells with the ferroptosis inducer erastin or sorafenib treatment (55). In the present study, loss of NUDCD1 activity attenuated GPX4 expression, elevated FTH1 expression, enhanced GSSG/GSH levels, and reduced GSH levels in HCC cells, demonstrating that NUDCD1 suppression facilitated the ferroptosis of HCC cells. The most ferroptosis-related genes were positively associated with HCC and exhibited prognostic value in HCC (56). SLC7A11, SLC1A5, RPL8, CARS1 and TFRC could serve as potential biomarkers for drug screening and provide additional targets for the immunotherapy of HCC (57). The present study suggests that the five ferroptosis-related genes could elucidate the molecular mechanisms of HCC and lead to a new direction for the improvement of predictive and preventive for HCC.
Several limitations should be pointed out. Although we screened 10 ferroptosis-related genes, more experiments will be implemented to verify their functions in the ferroptosis of HCC cells. Additionally, these ferroptosis-related genes were post-transcriptionally regulated by circRNA/miRNA and m1A/m5C/m6A RNA modifications, which should be experimentally validated in HCC.
Conclusions
Collectively, the present study determined 10 ferroptosis-related genes (UGT1A6, ATP6V1C1, MAFG, NUDCD1, PPP1R1A, TSKU, CTSB, AIFM2, CTSA, and CTNND2) in HCC, which were post-transcriptionally regulated by circRNA/miRNA and m1A/m5C/m6A RNA modifications. Among them, in vitro experiments demonstrated that suppression of NUDCD1 facilitated the ferroptosis of HCC cells. Altogether, our findings identified new and yet-unrecognized regulatory molecular mechanisms of ferroptosis in HCC.
Acknowledgments
The authors appreciate the academic support from the AME Hepatocellular Carcinoma Collaborative Group.
Funding: This work was funded by Nantong Municipal Science and Technology Project (Nos. JCZ20119, JCZ21063, JC12022108, MSZ20041) and scientific research special project of Nantong Health Commission (No. MS2022059)
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5750/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5750/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5750/coif). XW serves as an Editor-in-Chief of Annals of Translational Medicine from August 2019 to July 2024. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Lee TK, Guan XY, Ma S. Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications. Nat Rev Gastroenterol Hepatol 2022;19:26-44. [Crossref] [PubMed]
- Llovet JM, Castet F, Heikenwalder M, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022;19:151-72. [Crossref] [PubMed]
- Zhu AX, Kang YK, Yen CJ, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2019;20:282-96. [Crossref] [PubMed]
- Yau T, Park JW, Finn RS, et al. Nivolumab versus sorafenib in advanced hepatocellular carcinoma (CheckMate 459): a randomised, multicentre, open-label, phase 3 trial. Lancet Oncol 2022;23:77-90. [Crossref] [PubMed]
- Kaseb AO, Hasanov E, Cao HST, et al. Perioperative nivolumab monotherapy versus nivolumab plus ipilimumab in resectable hepatocellular carcinoma: a randomised, open-label, phase 2 trial. Lancet Gastroenterol Hepatol 2022;7:208-18. [Crossref] [PubMed]
- Niu X, Chen L, Li Y, et al. Ferroptosis, necroptosis, and pyroptosis in the tumor microenvironment: Perspectives for immunotherapy of SCLC. Semin Cancer Biol 2022;86:273-85. [Crossref] [PubMed]
- Chen J, Li X, Ge C, et al. The multifaceted role of ferroptosis in liver disease. Cell Death Differ 2022;29:467-80. [Crossref] [PubMed]
- Lyu N, Zeng Y, Kong Y, et al. Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann Transl Med 2021;9:675. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 2022;19:188-206. [Crossref] [PubMed]
- Xu Q, Zhou L, Yang G, et al. CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biol Int 2020;44:2344-56. [Crossref] [PubMed]
- Song P, Tayier S, Cai Z, et al. RNA methylation in mammalian development and cancer. Cell Biol Toxicol 2021;37:811-31. [Crossref] [PubMed]
- Chen H, Yao J, Bao R, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer 2021;20:29. [Crossref] [PubMed]
- Chen YT, Shen JY, Chen DP, et al. Identification of cross-talk between m6A and 5mC regulators associated with onco-immunogenic features and prognosis across 33 cancer types. J Hematol Oncol 2020;13:22. [Crossref] [PubMed]
- Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 2021;6:74. [Crossref] [PubMed]
- Fan Z, Yang G, Zhang W, et al. Hypoxia blocks ferroptosis of hepatocellular carcinoma via suppression of METTL14 triggered YTHDF2-dependent silencing of SLC7A11. J Cell Mol Med 2021;25:10197-212. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Vejnar CE, Zdobnov EM. MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic Acids Res 2012;40:11673-83. [Crossref] [PubMed]
- John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol 2004;2:e363. [Crossref] [PubMed]
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 2020;48:D127-31. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. [Crossref] [PubMed]
- Huang HY, Lin YC, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 2020;48:D148-54. [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. Erratum in: Nat Rev Mol Cell Biol 2018;19:808. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Stockwell BR, Jiang X, Gu W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol 2020;30:478-90. [Crossref] [PubMed]
- Zhang R, Pan T, Xiang Y, et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact Mater 2022;13:23-36. [Crossref] [PubMed]
- Liang JY, Wang DS, Lin HC, et al. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int J Biol Sci 2020;16:2430-41. [Crossref] [PubMed]
- Shen H, Wu H, Sun F, et al. A novel four-gene of iron metabolism-related and methylated for prognosis prediction of hepatocellular carcinoma. Bioengineered 2021;12:240-51. [Crossref] [PubMed]
- Yi K, Liu J, Rong Y, et al. Biological Functions and Prognostic Value of Ferroptosis-Related Genes in Bladder Cancer. Front Mol Biosci 2021;8:631152. [Crossref] [PubMed]
- Chen X, Yu C, Kang R, et al. Cellular degradation systems in ferroptosis. Cell Death Differ 2021;28:1135-48. [Crossref] [PubMed]
- Doll S, Freitas FP, Shah R, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019;575:693-8. [Crossref] [PubMed]
- Hu BB, Wang XY, Gu XY, et al. N6-methyladenosine (m6A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer 2019;18:178. [Crossref] [PubMed]
- Qu N, Qin S, Zhang X, et al. Multiple m6A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer 2020;20:165. [Crossref] [PubMed]
- Li D, Li K, Zhang W, et al. The m6A/m5C/m1A Regulated Gene Signature Predicts the Prognosis and Correlates With the Immune Status of Hepatocellular Carcinoma. Front Immunol 2022;13:918140. [Crossref] [PubMed]
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol 2021;18:280-96. [Crossref] [PubMed]
- Wang W, Green M, Choi JE, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. [Crossref] [PubMed]
- Lang X, Green MD, Wang W, et al. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov 2019;9:1673-85. [Crossref] [PubMed]
- Dai E, Han L, Liu J, et al. Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 2020;16:2069-83. [Crossref] [PubMed]
- Xu W, Rao Q, An Y, et al. Identification of biomarkers for Barcelona Clinic Liver Cancer staging and overall survival of patients with hepatocellular carcinoma. PLoS One 2018;13:e0202763. [Crossref] [PubMed]
- Shi C, Weng M, Zhu H, et al. NUDCD1 knockdown inhibits the proliferation, migration, and invasion of pancreatic cancer via the EMT process. Aging (Albany NY) 2021;13:18298-309. [Crossref] [PubMed]
- Han B, Xu K, Feng D, et al. miR-144 inhibits the IGF1R-ERK1/2 signaling pathway via NUDCD1 to suppress the proliferation and metastasis of colorectal cancer cells: a study based on bioinformatics and in vitro and in vivo verification. J Cancer Res Clin Oncol 2022;148:1903-18. [Crossref] [PubMed]
- He B, Xia S, Zhang Z. NudCD1 Promotes the Proliferation and Metastasis of Non-Small Cell Lung Cancer Cells through the Activation of IGF1R-ERK1/2. Pathobiology 2020;87:244-53. [Crossref] [PubMed]
- Wu Y, Zhang S, Gong X, et al. The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression. Mol Cancer 2020;19:39. [Crossref] [PubMed]
- Zhang Y, Swanda RV, Nie L, et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat Commun 2021;12:1589. [Crossref] [PubMed]
- Guerriero E, Capone F, Accardo M, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J Histochem 2015;59:2540. [Crossref] [PubMed]
- Chen Y, Li L, Lan J, et al. CRISPR screens uncover protective effect of PSTK as a regulator of chemotherapy-induced ferroptosis in hepatocellular carcinoma. Mol Cancer 2022;21:11. [Crossref] [PubMed]
- Wang Q, Bin C, Xue Q, et al. GSTZ1 sensitizes hepatocellular carcinoma cells to sorafenib-induced ferroptosis via inhibition of NRF2/GPX4 axis. Cell Death Dis 2021;12:426. [Crossref] [PubMed]
- Bai T, Lei P, Zhou H, et al. Sigma-1 receptor protects against ferroptosis in hepatocellular carcinoma cells. J Cell Mol Med 2019;23:7349-59. [Crossref] [PubMed]
- Tang B, Zhu J, Li J, et al. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun Signal 2020;18:174. [Crossref] [PubMed]
- Zhang B, Zhao J, Liu B, et al. Development and Validation of a Novel Ferroptosis-Related Gene Signature for Prognosis and Immunotherapy in Hepatocellular Carcinoma. Front Mol Biosci 2022;9:940575. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


