
Pan-cancer analysis of DEPDC1 as a candidate prognostic biomarker and associated with immune infiltration
Highlight box
Key findings
• DEPDC1 is abnormally upregulated in pan-carcinoma tissues, and its expression is significantly different in immune subtypes and molecular subtypes of various tumors, which also has excellent diagnostic and prognostic value. Its main functions are to regulate cell cycle, proliferation, DNA damage and repair, and also to inhibit immune infiltration.
What is known and what is new?
• DEPDC1 is a newly reported tumor-associated gene.
• DEPDC1 is overactivated and expressed in pan-cancer and is also significantly positively correlated with many immune checkpoints.
What is the implication, and what should change now?
• DEPDC1 can be used as a broad-spectrum biomarker for the diagnosis and therapeutic of pan-cancer.
Introduction
Cancer is the second leading cause of death globally after ischemic heart disease, but is predicted to become the leading cause of death in 2060 (1). In 2021, there were 1,898,160 new cases of cancer and approximately 610,000 deaths in the United States (2). Despite decades of tremendous progress in multiple oncological treatment strategies (3), the efficacy of currently available therapies remains poor. The past decade has witnessed dramatic advances in cancer treatment through immunotherapy (4). The human immune system is an important barrier for preventing the occurrence and development of tumors. Unfortunately, cancer cells have evolved multiple strategies to evade the immune system, which has slowed the development of cancer immunotherapy strategies (5-7). Identifying biomarkers with the potential to respond to immunotherapy and understanding the roles of these markers may guide the clinical development of immunotherapy strategies for patients with cancer (8).
DEPDC1 is encoded by DEP domain containing 1, which is located in the 1p31.3 region of human chromosome 1. DEPDC1 is a newly reported novel tumor-related gene that is upregulated in several malignancies and has been shown to contribute to the tumorigenesis of various tumors, such as lung (9-11), gastric (12), hepatocellular (13-15), bladder (16,17), breast (18), prostate (19) and colorectal cancer (CRC) (20). Interestingly, DEPDC1 has almost undetectable expression in the other normal human tissues, with the exception of the testes (17,21). Harada et al. demonstrated that DEPDC1 can directly interact with zinc finger protein ZNF224 to form a complex, thereby inhibiting the transcription of A20, resulting in the activation of anti-apoptotic pathways after nuclear factor kappa B (NFkB) translocation to the nucleus (16). Feng et al. showed that DEPDC1 is essential for acceleration of nasopharyngeal carcinoma cell motility and cycle progression (22). In addition, DEPDC1 promoted CRC proliferation, migration, and invasion through the regulation of SUZ12-mediated H3K27Me3 (20). However, the precise role of DEPDC1 in pan-cancer has not been reported, and the role of DEPDC1 in immune infiltration remains to be fully elucidated.
This current study investigated the expression of DEPDC1 in patients with pan-cancer and examined its association with patient prognosis. We further analyzed the association between DEPDC1 and the immune cell infiltration and immune inhibitor genes. The results herein provide novel insights into the functional role of DEPDC1 in pan-cancer, highlighting a potential mechanism whereby DEPDC1 influences the tumor microenvironment, as well as cancer immunotherapy. This project systematically studies the expression, mutation, prognosis, and function of DEPEC1 in human pan-cancers, providing a broad-spectrum biomarker for cancer diagnosis and treatment. We present the following article in accordance with the REMARK reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5598/rc).
Methods
Mutation profiles
The cBioPortal for cancer genomics (http://www.cbioportal.org) is an open-access repository of cancer genomics datasets (23,24). The cBioPortal database was used to analyze DEPDC1 genomic changes in order to analyze DEPDC1 mutations in pan-carcinoma.
Data collection
The RNA expression profiles and clinical data of 33 tumor types and 15,776 normal tissues samples were download from The Cancer Genome Atlas (TCGA) database and the Genotype-Tissue Expression (GTEx) database from the UCSC Xena database (http://xena.ucsc.edu/). To comprehensively investigated differential expression levels of DEPDC1 between tumor and adjacent normal tissues across TCGA cancer types, the Wilcoxon rank-sum test was performed, and P<0.05 was considered statistically significant (ns, P≥0.05; *P<0.05; **P<0.01; ***P<0.001) (25).
Protein-protein interaction networks
The STRING (https://string-preview.org/) database was used to analyze the protein interaction network of DEPDC1 and to obtain interacting genes and proteins.
Kyoto Encyclopedia of Genes and Genomes enrichment analysis and Gene Ontology
The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses and Gene Ontology (GO) was conducted for 50 DEPDC1-binding proteins using the ggplot2 package for visualization and the cluster Profiler package for statistical analysis.
Immune cell infiltration
TIMER is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types (26,27). TIMER 2.0 (http://timer.cistrome.org/) was used to analyze the correlation of DEPDC1 expression levels with the infiltration levels of 34 immune cell types associated with pan-cancer in the XCELL webtool (28). A P value <0.05 was considered statistically significant.
Analysis of the immune and molecular subtypes associated with DEPDC1 in pan-cancer
The TISIDB database, an integrated repository portal for tumor-immune system interactions (29) (http://cis.hku.hk/TISIDB/index.php), was used to explore the correlations between DEPDC1 expression and immune subtypes or molecular subtypes in pan-cancer. A P value <0.05 was considered statistically significant.
Analysis of the prognostic and diagnostic value of DEPDC1
Univariate Cox regression analyses of DEPDC1 and clinical characteristics were conducted to assess its prognostic value in the overall survival (OS), disease specific survival (DSS), and progress-free interval (PFI) of pan-cancer patients. The receiver operating characteristic (ROC) curve was used to assess the diagnostic value of DEPDC1 in pan-cancer.
Single-cell analysis
CancerSEA is the first dedicated database aimed at comprehensively decoding the functional status of cancer cells at a single-cell resolution. CancerSEA (30) (http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp) was used to explore DEPDC1 family-expressed functions.
Oral squamous cell carcinoma samples and immunohistochemical staining of DEPDC1
From 2018 to 2019, a total of 57 oral squamous cell carcinoma (OSCC) samples and 12 normal healthy oral tissue samples were collected at Xiangya Hospital (the affiliated hospital of Central South University) in Changsha, Hunan. The inclusion and exclusion criteria of the OSCC patients and the construction of tissue microarray have been detailed in our previous study (31). Immunohistochemistry (IHC) was performed, as previously described (31), to evaluate the expression level of the DEPDC1 protein, using an anti-DEPDC1 antibody (Abcam, ab197246, 1:300). Negative controls, positive controls, and phosphate buffered saline (PBS) blank controls were used to determine the efficacy of the IHC staining. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of School of Life Sciences, Central South University (No. 2022-1-44) and informed consent was taken from all the patients.
Statistical analyses
The R (v.3.6.3) software was used for data visualization. GraphPad 8.0 was used for statistical analysis of the data. The Wilcoxon rank-sum test or Student’s t-test was performed for comparisons between groups. Correlation analysis was conducted using Pearson or Spearman’s test as appropriate. Survival analysis was performed using univariate Cox regression, with P<0.05 being considered statistically significant.
Results
Analysis of DEPDC1 expression in pan-cancer
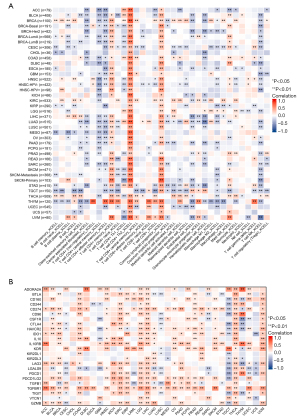
An unpaired analysis of DEPDC1 expression in pan-cancer revealed a significant upregulation of DEPDC1 in numerous cancers, including adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin sutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), thymoma (THYM), yterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS). In contrast, DEPDC1 was significantly downregulated in acute myeloid leukemia (LAML) and testicular germ cell tumors (TGCT) (Figure 1A). A paired analysis of DEPDC1 expression in pan-cancer showed that DEPDC1 was significantly upregulated in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, STAD, and UCSC, but did not change in PAAD, PRAD, READ, and THCA (Figure 1B). In addition, DEPDC1 expression was highest in TCGA tumor tissues in ESCA, CESC, and SARC (Figure 1C). However, the expression of DEPDC1 was at a lower level in all other normal tissues, except in the normal tissues of LAML and TGCT (Figure 1D).
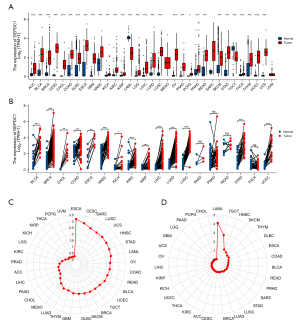
Correlations between DEPDC1 and immune subtypes of pan-cancer
Differentiating immune subtypes of tumors is of great clinical value for precision medicine of tumors (32). To determine the correlation of DEPDC1 gene expression with immune subtypes in pan-cancer, we explored the differential DEPDC1 expression in pan-cancer immune subtypes, namely, C1: wound healing; C2: interferon gamma (IFN-g) dominant; C3: inflammatory; C4: lymphocyte depleted; C5: immunologically quiet; and C6: tumor growth factor beta (TGF-b) dominant, from the TISIDB database. The outcomes of immune subtypes showed that DEPDC1 was lowest in C3 subtypes of BLCA (Figure 2A), BRCA (Figure 2B), COAD (Figure 2C), ESCA (Figure 2D), LIHC (Figure 2E), LUAD (Figure 2F), LUSC (Figure 2G), MESO (Figure 2H), OV (Figure 2I), PAAD (Figure 2J), PRAD (Figure 2K), READ (Figure 2L), SARC (Figure 2M), SKCM (Figure 2N), STAD (Figure 2O), TGCT (Figure 2P), THCA (Figure 2Q), and UCEC (Figure 2R). In addition, the expression of DEPDC1 was the lowest in the C5 immune subtypes of tumors including ACC (Figure 2S), GBM (Figure 2T), KIRC (Figure 2U), KIRP (Figure 2V), and LGG (Figure 2W).
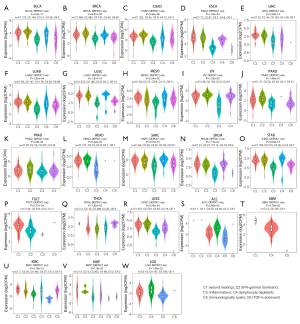
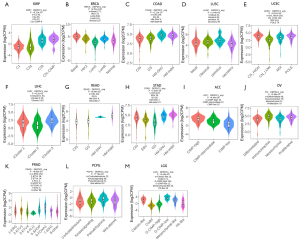
Correlations between DEPDC1 and molecular subtypes of pan-cancer
Meanwhile, significant differences exist in the expression of DEPDC1 in different molecular subtypes. The results indicated that the expression of DEPDC1 in C2c-CpG island methylator phenotype (CIMP) is highest in KIRP relative to other molecular subtypes (Figure 3A). In BRCA, DEPDC1 was highest expressed in the molecular subtype of basal (Figure 3B). In COAD, DEPDC1 was highest expressed in the molecular subtype of Hypermutated- single nucleotide variants (HM-SNV) (Figure 3C). In LUSC, DEPDC1 was highest expressed in the molecular subtype of primitive (Figure 3D). In UCEC, DEPDC1 was highest expressed in the molecular subtype of copy number high (CN_HIGH) (Figure 3E). In LIHC, DEPDC1 was highest expressed in the molecular subtype of iCluster.2 (Figure 3F). In READ, DEPDC1 was highest expressed in the molecular subtype of Hypermutated (HM)-indel (Figure 3G). In STAD, DEPDC1 was highest expressed in the molecular subtype of HM-indel (Figure 3H). In ACC, DEPDC1 was highest expressed in the molecular subtype of CIMP-high (Figure 3I). In OV, DEPDC1 was highest expressed in the molecular subtype of immunoreactive (Figure 3J). In PRAD, DEPDC1 was highest expressed in the molecular subtype of 6-FOXA1 (Figure 3K). In PCPG, DEPDC1 was highest expressed in the molecular subtype of Wnt-altered (Figure 3L). In LGG, DEPDC1 was highest expressed in the molecular subtype of G-CIMP-low (Figure 3M).
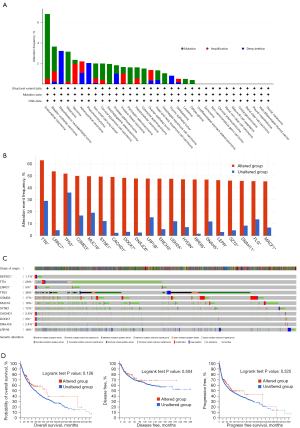
The landscape of DEPDC1 mutation profile in different tissues
The cBioPortal was used to examine the mutation frequency of DEPDC1 in the TCGA pan-cancer atlas (10,967 samples in 32 studies). Endometrial carcinoma was found to have the highest mutation level with the DEPDC1 alteration frequency exceeding 7% (Figure 4A). In addition, the frequency and pattern of genetic alterations in numerous genes was similar to that of DEPDC1 genetic alterations (Figure 4B,4C), including titin (TTN), leucine rich repeat containing 7 (LRRC7), tumor protein p53 (TP53), CUB and Sushi multiple domains 2 (CSMD), mucin 16, cell surface associated (MUC16), spectrin repeat containing nuclear envelope protein 1 (SYNE1), cache domain containing 1 (CACHD1), dedicator of cytokinesis 7 (DOCK7), DnaJ heat shock protein family (Hsp40) member C6 (DNAJC6), and LDL receptor related protein 1B (LRP1B). However, DEPDC1 genetic alterations in the study cohort were not associated with prognostic outcomes (Figure 4D).
Diagnostic value of DEPDC1 in pan-cancer
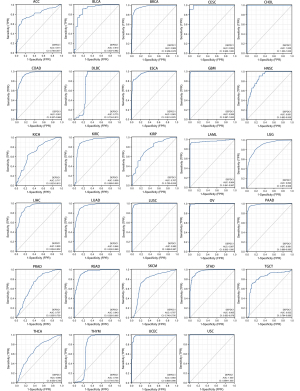
A ROC analysis of DEPDC1 in pan-cancer was performed to assess its diagnostic value. The results revealed that DEPDC1 has diagnostic value in pan-cancer (Figure 5), with area under the curve (AUC values) as follows: ACC, AUC =0.811; BLCA, AUC =0.910; BRCA, AUC =0.969; CESC, AUC =0.993; CHOL, AUC =1.000; COAD, AUC =0.951; DLBC, AUC =0.773; ESCA, AUC =0.963; GBM, AUC =0.999; HNSC, AUC =0.891; KICH, AUC =0.717; KIRC, AUC =0.909; KIRP, AUC =0.767; LAML, AUC =0.964; LGG, AUC =0.893; LIHC, AUC =0.928; LUAD AUC =0.946; LUSC, AUC =0.992; OV, AUC =0.997; PAAD, AUC =0.981; PRAD, AUC =0.707; READ, AUC =0.943; SKCM, AUC =0.767; STAD, AUC =0.965; TGCT, AUC =0.832; THCA, AUC =0.690; THYM, AUC =0.754; UCEC, AUC =0.983; and UCS, AUC =1.000. Among them, the prediction accuracy of DEPDC1 for CESC, CHOL, LUSC, OV, and UCS was greater than 0.99.
Analysis of the prognostic value of DEPDC1 in pan-cancer
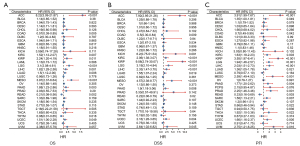
To investigate whether DEPDC1 has potential prognostic value, univariate Cox regression was used to explore the relationship between DEPDC1 expression and prognosis in multiple tumor types. High DEPDC1 expression was associated with poor OS in patients with [hazard ratio (HR) =8.22; 95% confidence interval (CI): 2.09–21.87, P<0.001], KIRC (HR =1.52; 95% CI: 1.12–2.07, P=0.007), KIRP (HR= 2.23; 95% CI: 1.20–3.19, P=0.011), LGG (HR =2.1; 95% CI: 2.08–3.61, P<0.001), LIHC (HR =1.82; 95% CI: 1.28–2.58, P=0.001), LUAD (HR =1.5; 95% CI: 1.12–2.00, P=0.006), MESO (HR =2.97; 95% CI: 2.27–6.63, P<0.001), PAAD (HR =1.88; 95% CI: 1.22–2.86, P=0.002), and UCEC (HR =1.51; 95% CI: 1.00–2.28, P=0.039). Higher DEPDC1 expression was associated with good OS in cases of COAD [HR =0.57 (0.28–0.85), P=0.005], READ (HR =0.21; 95% CI: 0.08–0.56, P=0.002), and THYM (HR =0.08; 95% CI: 0.01–0.65, P=0.018) (Figure 6A). In addition, DSS analysis showed that high DEPDC1 expression was a marker for poor outcome for patients with ACC (HR= 7.92; 95% CI: 2.95–21.26, P<0.001), KIRC (HR =2.06; 95% CI: 1.38–3.07, P<0.001), KIRP (HR= 9.08; 95% CI: 2.74–30.07, P<0.001), LGG (HR =3.19; 95% CI: 2.10–4.84, P<0.001), LIHC (HR =2.33; 95% CI: 1.47–3.70, P=0.001), LUAD (HR =1.55; 95% CI: 1.07–2.24, P=0.019), MESO (HR =4.57; 95% CI: 2.38–8.77, P<0.001), PAAD (HR =1.9; 95% CI: 1.18–3.06), P=0.008), UCEC (HR =2.16; 95% CI: 1.28–3.65, P=0.004), and UVM (HR =2.64; 95% CI: 1.02–6.82, P=0.045). Higher DEPDC1 expression was associated with good OS in cases of COAD (HR =0.5; 95% CI: 0.30–0.84, P=0.009) and READ (HR =0.22; 95% CI: 0.06–0.79, P=0.02) (Figure 6B). Furthermore, PFI analysis showed that high DEPDC1 expression was a marker for poor outcome for patients with ACC (HR= 3.91; 95% CI: 1.99–7.67, P<0.001), BLCA; CI (HR =1.35; 95% CI: 1.00–1.81, P<0.049), KIRC (HR =1.63; 95% CI: 1.18–2.23, P=0.003), KIRP (HR= 3.55; 95% CI: 1.97–6.41, P<0.001), LGG (HR =1.94; 95% CI: 1.46–2.57, P<0.001), LIHC (HR =2.03; 95% CI: 1.51–2.73, P=0.001), LUAD (HR =1.36; 95% CI: 1.05–1.78, P=0.021), MESO (HR =2.59; 95% CI: 1.49–4.52, P=0.001), PAAD (HR =1.77; 95% CI: 1.20–2.62, P=0.004), PRAD (HR =2.25; 95% CI: 1.46–3.46, P=0.001), SARC (HR =1.58; 95% CI: 1.13–2.21, P=0.007), THCA (HR =1.92; 95% CI: 1.10–3.36, P=0.022), UCEC (HR =1.66; 95% CI: 1.16–2.37, P=0.005) and UVM (HR =2.75; 95% CI: 1.21–6.25, P=0.016). Higher DEPDC1 expression was associated with good PFI in cases of COAD (HR =0.7; 95% CI: 0.49–0.99, P=0.042), READ (HR =0.33; 95% CI: 0.16–0.69, P=0.003), and STAD (HR =0.66; 95% CI: 0.46–0.94, P=0.022) (Figure 6C). These results indicated that increased DEPDC1 expression was associated with poor prognosis in a variety of tumor types, but in a few tumors they are associated with a good prognosis.
PPI network and GO and KEGG enrichment analyses
To investigate the functional mechanism of DEPDC1 in carcinogenesis, we used the following parameters as screening conditions in the STRING (https://www.string-db.org/) database: active interaction sources (“Textmining, Experiments, Databases, Co-expression, Neighborhood, Gene Fusion, and Co-occurrence”) and minimum required interaction score ["medium confidence (0.400)"] yielded the top 50 proteins interacting with DEPDC1. The results showed that the genes most associated with DEPDC1 were kinesin family member 20A (KIF20A), DLG associated protein 5 (DLGAP5), centromere protein F (CENPF), Rho GTPase activating protein 11A (ARHGAP11A), assembly factor for spindle microtubules (ASPM), kinesin family member 11 (KIF11), TTK protein kinase (TTK), BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B), DNA topoisomerase II alpha (TOP2A), and zinc finger protein 224 (ZNF224) (Figure 7A). GO and KEGG pathway enrichment analyses of the 50 targeted binding proteins were obtained using the STRING tool. The results revealed that the proteins of interest were mainly involved in cell cycle, oncyte meiosis, progesterone-mediated oncyte maturation, cellular senescence, p53 signaling pathway, human immunodeficiency virus 1 infection, viral carcinogenesis, and FoxO signaling pathway. The main biological processes (BPs) were mitotic nuclear division, nuclear division, organelle fission, mitotic sister chromatid segregation, and sister c chromatid segregation. The main molecular functions (MFs) were microtubule binding, histone kinase activity, tubulin binding, chemokine activity, and motor activity. These results support the conclusion that DEPDC1 is essential for tight junctions, cell cycle and nuclear division.
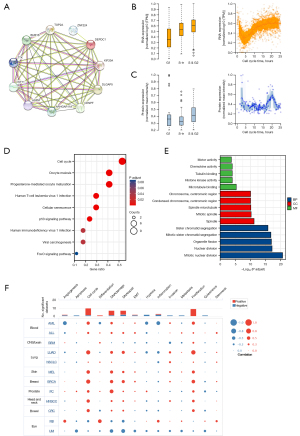
To determine whether the changes in DEPDC1 expression at the protein and transcript level were associated with the cell cycle, single-cell RNA-sequencing data from the human protein atlas (HPA) (https://www.proteinatlas.org/) (33) was analyzed. Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) revealed increased DEPDC1 RNA and protein in U-2 OS cells expression in relation to cell cycle progression (Figure 7B,7C). To investigate the functions of the DEPDC1 family in pan-cancer, we performed single-cell analysis using CancerSEA. The results indicated that DEPDC1 positively regulated cell cycle, DNA damage, DNA repair, and proliferation in blood cancer, lung cancer, skin cancer, breast cancer, prostate cancer, HNSCC, and bowel cancer. These results suggested that DEPDC1 is essential for cell cycle progression and proliferation of tumor cells (Figure 7D-7F).
Immune cell infiltration analyses
Immune infiltration of tumors is the frontier direction of current tumor research, and thus, identifying valuable biomarkers of tumor immune infiltration is crucial. To determine whether DEPDC1 could serve as a pan-cancer marker of immune infiltration, we explored the association of DEPDC1 with various immune cells in human cancers using XCELL metrics in TIMER2.0 (Figure 8A). The results showed that the expression level of DEPDC1 was positively correlated with CD4+ T helper 2 (Th2) cells and common lymphoid progenitors. Correspondingly, there was a negative correlation between the expression level of DEPDC1 and natural killer (NK) T cells, CD4+ central memory T cells and CD4+ effector memory T cells. In most cancers, except TGCT, THYM, and UCS, robust and significant relationships existed between DEPDC1 expression and expression levels of recognized immune checkpoints including interleukin 10 receptor subunit beta (IL10RB), CD274, transforming growth factor beta receptor 1 (TGFBR1), kinase insert domain receptor (KDR), and programmed cell death 1 ligand 2 (PDCD1LG2). This suggested a potential synergy of DEPDC1 with known immune checkpoints (Figure 8B).
DEPDC1 is upregulated in OSCC and has prognostic value
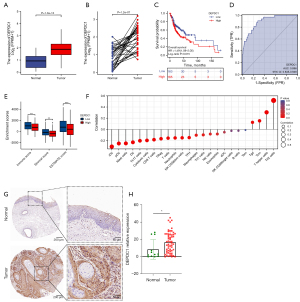
It is well-documented that DEPDC1 is upregulated in a variety of tumors and is associated with poor prognosis. To verify this, we analyzed the expression of DEPDC1 and its prognostic value in OSCC. As shown in Figure 9A,9B, both paired and unpaired analyses in TCGA-OSCC showed that the mRNA expression level of DEPDC1 was significantly upregulated. DEPDC1 was also associated with poor prognosis in OSCC (Figure 9C), and the expression of DEPDC1 in OSCC can be used to differentiate normal and tumor tissues (Figure 9D). Tumor samples with low expression of DEPDC1 in OSCC had higher immune score, stromal score, and ESTMATE score, indicating that DEPDC1 inhibited immune infiltration in OSCC (Figure 9E). In addition, using the single sample gene set enrichment analysis (ssGSEA) method, we found that DEPDC1 was significantly positively correlated with Th2 cells and T helper cells, and significantly negatively correlated with presenting cells, which is consistent with our analysis in pan-cancer (Figure 9F). To further confirm the abnormally high expression of DEPDC1 in OSCC, we collected 57 OSCC samples and 12 normal samples, and analyzed the expression of DEPDC1 after IHC staining of these samples. The results showed that DEPDC1 was indeed significantly upregulated in OSCC (Figure 9G-9H).
Discussion
Cancer presents a global threat to health and screening for valuable diagnostic and therapeutic biomarkers is crucial. DEPDC1 is a highly evolutionarily conserved gene that is widely expressed in animals and plants in nature (18,21). Current studies have shown that DEPDC1 has orthologs in 304 organisms and humans (13,16,34). In humans, the DEPDC1 gene is a novel oncogene located on chromosome 1 (21). At present, research related to the function of DEPDC1 is still in its infancy. Furthermore, the role of DEPDC1 in pan-cancer has not been reported. This project evaluated DEPDC1 expression changes in pan-cancer. Analysis of the sequencing data of 33 tumors and corresponding normal samples obtained from the TCGA and GTEx databases revealed that DEPDC1 was significantly upregulated in 29 human cancers, including ACC, BRCA, BLCA, CESC, COAD, CHOL, DLBC, ESCA, GBM, HNSC, KIRP, KIRC, KICH, LUSC, LUAD, LIHC, LGG, OV, PCPG, PRAD, PAAD, READ, SKCM, STAD, SARC, THYM, THCA, UCS, and UCEC. Meanwhile, DEPDC1 was significantly downregulated in LAML and TGCT. This finding suggested that DEPDC1 is abnormally amplified in most malignancies and acts as a proto-oncogene and may be involved in tumorigenesis or cancer development.
The determination of the immune subtype of tumors is the basis for personalized and precise treatment of tumors. Our results showed that DEPDC1 is significantly differentially expressed in different immune subtypes of 23 cancers, including ACC, BLCA, BRCA, COAD, ESCA, GBM, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, and UCEC. It is worth noting that DEPDC1 expression was the lowest in the C3 and C5 subtypes. Immunological landscape studies on cancer have shown that C3 (inflammatory subtype) and C5 (immune quiet subtype) have better survival outcomes than other immune subtypes (35). These results suggested that DEPDC1 as a proto-oncogene is associated with a poor prognosis. At the same time, DEPDC1 expression levels were also significantly different in molecular subtypes of 13 cancer types, including KIRP, BRCA, LUSC, COAD, UCEC, LIHC, READ, ACC, OV, PRAD, PCPG, LGG, and STAD. This suggested that DEPDC1 may be a key indicator of tumor immune subtype or molecular typing. Therefore, taking the immune subtype or molecular subtype of the tumor as a starting point may help to understand the role of DEPDC1 in pan-cancer.
To evaluate the value of DEPDC1 as a diagnostic and prognostic marker in pan-cancer, we performed ROC curve analysis on pan-cancer and found that DEPDC1 can effectively distinguish normal and tumor samples in all types of cancers (AUC >0.7), especially in CESC, CHOL, LUSC, OV, and UCS (AUC >0.99). Furthermore, pan-cancer survival analysis revealed that DEPDC1 was significant in OS, DSS, and PFI in ACC, COAD, KIRP, KIRC, LGG, LUAD, LIHC, MESO, PAAD, READ, THYM, and UCEC. These results suggested that DEPDC1 has an extremely important diagnostic value in pan-cancer and can be used as a prognostic marker in specific cancers. Therefore, DEPDC1 can be used as a potential biomarker or therapeutic target for precision medicine of tumors in the future.
To gain an in-depth understanding of the biological function of DEPDC1, we obtained 50 interacting proteins of DEPDC1 through the STRING database for GO and KEGG pathway enrichment analysis. The analysis revealed that DEPDC1 is mainly involved in cell cycle, cell meiosis, and progesterone-mediated cell cycle, as well as maturation, cellular senescence, p53 signaling, human immunodeficiency virus 1 infection, viral carcinogenesis, and FoxO signaling. The associated BPs were primarily involved in the mitotic nuclear division, nuclear division, organelle fission, mitotic sister chromatid segregation, and sister c chromatid segregation. The main enriched MFs were microtubule binding, histone kinase activity, tubulin binding, chemokine activity, and motor activity. These results suggested that DEPDC1 is essential for cell cycle regulation and regulates nuclear division in cells. This is consistent with the results of existing functional studies related to DEPDC1 (12,22,36).
In addition to the involvement of DEPDC1 in regulating the cell cycle, the results herein demonstrated that DEPDC1 expression is highly correlated with immune infiltration. Through the TIMER2.0 database, we found that DEPDC1 was significantly associated with the abundance of 35 XCELL immune infiltrating cells in pan-cancer. In particular, DEPDC1 was a significant positive correlation with CD4+ Th2 cells and common lymphoid progenitor cells. Conversely, DEPDC1 was significantly negatively correlated with NK cells, CD4+ central memory T cells, and CD4+ effector memory T cell. These results suggested that DEPDC1 can activate tumor immune escape through the combined action of activating Th2 cells and inhibiting NK cells, thereby enabling tumor cells to obtain a more stable environment to deteriorate the body’s health.
Cancer patients usually do not have a significant reduction in T cell numbers, but most of the T cell function is lost, which is often referred to as T cell exhaustion (37). T cell exhaustion is usually accompanied by enhanced PD-1 expression on the surface of T cells, and a large amount of PDL1 and PDL2 expressed on the surface of the corresponding tumor cells. We found that DEPDC1 positively correlated with T cell exhaustion marker genes such as CD274, TGFBR1, KDR, PDCD1LG2, GZMB, and LAG2 in pan-cancer. This suggested that DEPDC1 may be involved in suppressing tumor immunity by interacting with immune checkpoints.
There were some limitations to this study. This investigation used public databases including TCGA to explore the relationship between DEPDC1 and pan-cancer. Although clinical samples from our hospital were used for verification, further clinical data should be analyzed to confirm these results. The biological functions associated with DEPDC1 were determined by bioinformatics, and future experiments involving in vitro and in vivo experiments are warranted.
Conclusions
In conclusion, the results herein revealed that DEPDC1 is up-regulated in pan-cancer tissues, with the exception of LAML and TGCT, and plays an immune-oncogenic role in pan-cancer. Indeed, DEPDC1 can be used as a broad-spectrum biomarker for the diagnosis and therapeutic of pan-cancer.
Acknowledgments
We thank Professor Tong Su (Department of Oral and Maxillofacial Surgery, Center of Stomatology, Xiangya Hospital of Central South University, Changsha, China) and Professor Kun Xia (Center for Medical Genetics & Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, China) for their help in the research direction and experimental design.
Funding: The study was supported by Health Commission of Zhejiang Province (No. 2019KY151).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5598/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5598/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5598/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of School of Life Sciences, Central South University (No. 2022-1-44) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mattiuzzi C, Lippi G. Current Cancer Epidemiology. J Epidemiol Glob Health 2019;9:217-22. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 2018;15:95-111. [Crossref] [PubMed]
- Weiss SA, Wolchok JD, Sznol M. Immunotherapy of Melanoma: Facts and Hopes. Clin Cancer Res 2019;25:5191-201. [Crossref] [PubMed]
- Zhou X, Du J, Liu C, et al. A Pan-Cancer Analysis of CD161, a Potential New Immune Checkpoint. Front Immunol 2021;12:688215. [Crossref] [PubMed]
- Zeng D, Ye Z, Wu J, et al. Macrophage correlates with immunophenotype and predicts anti-PD-L1 response of urothelial cancer. Theranostics 2020;10:7002-14. [Crossref] [PubMed]
- Cao R, Yang F, Ma SC, et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020;10:11080-91. [Crossref] [PubMed]
- Emens LA. Breast Cancer Immunotherapy: Facts and Hopes. Clin Cancer Res 2018;24:511-20. [Crossref] [PubMed]
- Shen J, Xi M. DEPDC1 is Highly Expressed in Lung Adenocarcinoma and Promotes Tumor Cell Proliferation. Zhongguo Fei Ai Za Zhi 2021;24:453-60. [PubMed]
- Wang Q, Li A, Jin J, et al. Targeted interfering DEP domain containing 1 protein induces apoptosis in A549 lung adenocarcinoma cells through the NF-κB signaling pathway. Onco Targets Ther 2017;10:4443-54. [Crossref] [PubMed]
- Liu C, Li X, Hao Y, et al. STAT1-induced upregulation of lncRNA KTN1-AS1 predicts poor prognosis and facilitates non-small cell lung cancer progression via miR-23b/DEPDC1 axis. Aging (Albany NY) 2020;12:8680-701. [Crossref] [PubMed]
- Gong Z, Chu H, Chen J, et al. DEPDC1 upregulation promotes cell proliferation and predicts poor prognosis in patients with gastric cancer. Cancer Biomark 2021;30:299-307. [Crossref] [PubMed]
- Zhou C, Wang P, Tu M, et al. DEPDC1 promotes cell proliferation and suppresses sensitivity to chemotherapy in human hepatocellular carcinoma. Biosci Rep 2019;39:BSR20190946. [Crossref] [PubMed]
- Amisaki M, Yagyu T, Uchinaka EI, et al. Prognostic Value of DEPDC1 Expression in Tumor and Non-tumor Tissue of Patients With Hepatocellular Carcinoma. Anticancer Res 2019;39:4423-30. [Crossref] [PubMed]
- Guo W, Li H, Liu H, et al. DEPDC1 drives hepatocellular carcinoma cell proliferation, invasion and angiogenesis by regulating the CCL20/CCR6 signaling pathway. Oncol Rep 2019;42:1075-89. [Crossref] [PubMed]
- Harada Y, Kanehira M, Fujisawa Y, et al. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res 2010;70:5829-39. [Crossref] [PubMed]
- Kanehira M, Harada Y, Takata R, et al. Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene 2007;26:6448-55. [Crossref] [PubMed]
- Zhao H, Yu M, Sui L, et al. High Expression of DEPDC1 Promotes Malignant Phenotypes of Breast Cancer Cells and Predicts Poor Prognosis in Patients With Breast Cancer. Front Oncol 2019;9:262. [Crossref] [PubMed]
- Huang L, Chen K, Cai ZP, et al. DEPDC1 promotes cell proliferation and tumor growth via activation of E2F signaling in prostate cancer. Biochem Biophys Res Commun 2017;490:707-12. [Crossref] [PubMed]
- Wang Q, Jiang S, Liu J, et al. DEP Domain Containing 1 Promotes Proliferation, Invasion, and Epithelial-Mesenchymal Transition in Colorectal Cancer by Enhancing Expression of Suppressor of Zest 12. Cancer Biother Radiopharm 2021;36:36-44. [Crossref] [PubMed]
- Kharrat A, Millevoi S, Baraldi E, et al. Conformational stability studies of the pleckstrin DEP domain: definition of the domain boundaries. Biochim Biophys Acta 1998;1385:157-64. [Crossref] [PubMed]
- Feng X, Zhang C, Zhu L, et al. DEPDC1 is required for cell cycle progression and motility in nasopharyngeal carcinoma. Oncotarget 2017;8:63605-19. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol 2017;35:314-6. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [Crossref] [PubMed]
- Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res 2019;47:D900-8. [Crossref] [PubMed]
- Hu X, Xia K, Xiong H, et al. G3BP1 may serve as a potential biomarker of proliferation, apoptosis, and prognosis in oral squamous cell carcinoma. J Oral Pathol Med 2021;50:995-1004. [Crossref] [PubMed]
- Chen Y, Sun Z, Chen W, et al. The Immune Subtypes and Landscape of Gastric Cancer and to Predict Based on the Whole-Slide Images Using Deep Learning. Front Immunol 2021;12:685992. [Crossref] [PubMed]
- Colwill K. Gräslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods 2011;8:551-8. [Crossref] [PubMed]
- Ramalho-Carvalho J, Martins JB, Cekaite L, et al. Epigenetic disruption of miR-130a promotes prostate cancer by targeting SEC23B and DEPDC1. Cancer Lett 2017;385:150-9. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. [Crossref] [PubMed]
- Mi Y, Zhang C, Bu Y, et al. DEPDC1 is a novel cell cycle related gene that regulates mitotic progression. BMB Rep 2015;48:413-8. [Crossref] [PubMed]
- Jiang W, He Y, He W, et al. Exhausted CD8+T Cells in the Tumor Immune Microenvironment: New Pathways to Therapy. Front Immunol 2020;11:622509. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


