Differential expression of microRNAs in bile duct obstruction-induced liver fibrosis and the identification of a novel liver fibrosis marker miR-1295b-3p
Highlight box
Key findings
• Differentially expressed microRNAs were found in bile duct obstruction-induced liver fibrosis, and overexpression of miR-1295b-3p may be a potential treatment for liver fibrosis.
What is known and what is new?
• The obstruction of intrahepatic or extra hepatic bile ducts causes cholestasis liver fibrosis. MicroRNA can participate in the regulation of liver fibrosis.
• This study identified potential target genes and signal pathways that are related to the differentially expressed microRNAs in bile duct obstruction-induced liver fibrosis, and overexpressing miR-1295b-3p may be a potential treatment for liver fibrosis.
What is the implication, and what should change now?
• The differentially expressed microRNAs may be novel biological markers for the diagnosis, treatment, and prognosis of bile duct obstruction-induced liver fibrosis. The specific regulatory mechanisms of microRNAs in bile duct obstructive liver fibrosis warrant further research.
Introduction
Bile duct obstruction-induced liver fibrosis is a type of liver fibrosis mainly caused by cholestatic liver injury. It is prevalent in common bile duct stones, ampullary tumors, and cholangiocarcinomas. These diseases cause bile to accumulate in the liver, further stimulating liver cell inflammation and damaging the liver structure, causing liver fibrosis (1). Regardless of the cause, the recognized liver fibrosis mechanism is that chronic liver damage produces various pro-fibrotic mediators, such as excessive reactive oxygen species (ROS), transforming growth factor β (TGF-β), and cytokines, thus activating hepatic stellate cells (HSCs) and accelerating cell proliferation. As a result, the levels of alpha smooth muscle actin (α-SMA) and collagen increase significantly (2), accelerating liver fibrosis. Prolonged and increased damage of the liver and the deposition of extracellular matrix (ECM) exacerbates liver fibrosis, thereby reducing the blood flow to the liver cell plate and increasing liver hypoxia and liver cell death (3). The primary treatment for this type of cholestatic disease is surgery. Bile duct puncture and drainage are the most direct methods to relieve obstruction and drain the bile. However, it is difficult for patients to recover from chronic liver damage caused by primary diseases, which may further develop into cirrhosis or liver failure.
MicroRNAs are endogenous, non-coding small RNA molecules, with a length of approximately 20–24 nucleotides, which are highly conserved in evolution. MicroRNAs play an important regulatory role in various physiological processes, such as cell proliferation, apoptosis, and fat metabolism. Recent studies have shown that microRNAs affect liver fibrosis via various metabolic pathways. For instance, miRNA-29a inhibits BRD4 and reduces hepatic fibrosis in murine tissues by suppressing the activation of HSC, and miR-199a-3p exacerbates hepatic fibrosis by inhibiting the expression of caveolin-2 and triggering the TGF-β pathway (4,5). However, the important microRNA related to liver fibrosis has not been revealed to a great extent.
This study used gene sequencing and edgeR software to analyze human bile duct obstruction-induced liver fibrosis tissues and identify the differentially expressed microRNAs. Bioinformatics analysis was performed to predict the target genes involved in regulating signaling pathways to explore their roles in bile duct obstruction-induced liver fibrosis. Furthermore, we found that miR-1295b-3p is downregulated in LX-2 cells, and overexpression of miR-1295b-3p could inhibit liver fibrosis, leading to inhibition of proliferation and the induction of apoptosis in LX-2 cell. This study revealed the therapeutic potential of miR-1295b-3p in liver fibrosis, however, the specific targeting mechanisms warrant further investigation. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6416/rc).
Methods
Patient samples
A total of 8 pairs of liver fibrosis and matched normal liver tissues were screened. The liver fibrosis tissues samples were obtained from the patients with bile duct obstruction-induced liver fibrosis and the normal liver tissues were obtained from peri-hepatic-hemangioma liver tissues, who underwent liver resection from Department of Hepatobiliary-Pancreatic Surgery, Qingdao Municipal Hospital, between December 2017 to July 2021. The tissues were rapidly preserved at −80 ℃ or in 10% neutral-buffered formalin. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Qingdao Municipal Hospital (2020 No. 098) and informed consent was taken from all the patients.
Hematoxylin and eosin (HE) staining and Masson staining
Histological examination confirmed the liver fibrosis diagnosis. The liver tissue was fixed in 4% paraformaldehyde for 24 h, dehydrated, transparentized, embedded in paraffin, and sliced into 4 µm sections. The tissue structure was observed by HE staining and the collagen deposition was detected by Masson staining.
Screening of differentially expressed microRNAs
Normal liver tissues and bile duct obstruction-induced liver fibrosis tissues, three sets each, were analyzed using high-throughput sequencing (Beijing Berry Genomics Co., Ltd.) to detect the total microRNAs. EdgeR software was used to screen the differentially expressed microRNAs.
Target gene prediction and related analysis
Potential target genes of the identified microRNAs were predicted using the TargetScan7.1 and miRBase websites, and the miRanda and RNAhybrid software. The topGO software was used to perform the target Gene Ontology (GO) function enrichment analysis based on GO database. The KOBAS software was used to analyze the biochemical metabolic pathways and signal transduction pathways based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (DAVID; version 6.7, https://david.ncifcrf.gov).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Normal liver tissues and bile duct obstruction-induced liver fibrosis tissues (n=5) were analyzed using qRT-PCR. Total RNA was purified from liver tissues using TRIzol reagent (Tiangen, Beijing, China). For microRNA, reverse transcription was performed using a miRcute microRNA cDNA first strand synthesis kit (Funeng, Guangzhou, China). The comparative 2−ΔΔCt method was used to calculate the relative values of each gene. The primers designed for qRT-PCR are shown in Table 1.
Table 1
| Gene | Primer sequences | |
|---|---|---|
| Forward primer | Reverse primer | |
| miR-1295b-3p | 5'-GATCTGCGGCCTAATCAC-3' | 5'-CCAGATCCGTGGCCTATT-3' |
| Caspase-3 | 5'-ATGGAGAACAACAAAACCTCAGT-3' | 5'-TTGCTCCCATGTATGGTCTTTAC-3' |
| α-SMA | 5'-TCATGGTCGGTATGGGTCAG-3' | 5'-CCGTGCTCGATAGGGTACTT-3' |
| Bcl-2 | 5'-GGTGGGGTCATGTGTGTGG-3' | 5'-CGGTTCAGGTACTCAGTCATCC-3' |
| Bax | F:5'-CCCGAGAGGTCTTTTTCCGAG-3' | R:5'-CCAGCCCATGATGGTTCTGAT-3' |
| U6 | F:5'-GCTTCGGCAGCACATATACTAAAAT-3' | R:5'-CGCTTCACGAATTTGCGTGTCAT-3' |
| β-actin | 5'-TACCTGAAGCCCCAACTACAAA-3' | 5'-GTGCCCTGCCACATGATAAA-3' |
α-SMA, α-smooth muscle actin; qRT-PCR, quantitative real-time polymerase chain reaction.
Cell culture and transient transfection
The human liver LX-2 cell line was purchased from BeNa Culture Collection (Beijing, China) and cultivated in Dulbecco’s Minimum Essential Media (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS; Invitrogen) at 37 ℃, saturated humidity, and 5% CO2. Cells were treated with TGF-β1 (10 ng/mL) (Thermo Fisher Scientific) for 48 h. The miR-1295b-3p agomir and negative control (miR-NC) were obtained from Gema (Shanghai, China) and transfected into LX-2 cells using Lipofectamine 2000 reagent (Invitrogen). Let them stand at room temperature for 5 minutes, and add them to the corresponding wells of 6-well plate. After 6 h of transfection, the original culture medium was replaced for further culture. Afterward, cells were incubated for 48 h and harvested for further analysis.
Cell viability assay
The Cell Counting Kit 8 (CCK-8) assay (Dojindo) was used to evaluate the viability of LX-2 cells in accordance with the manufacturer’s instructions. Absorbance was analyzed at 450 nm using the microplate reader.
Western blot assay
The total proteome was collected from samples using RIPA lysis buffer (Aspen Biotechnology) containing protease inhibitors. The BCA assay kit (Aspen Biotechnology) was used to quantify proteins. The following primary antibodies were used to incubate the membranes: β-arrestin1 (ARRB1) (1:1,000, Abcam, UK), caspase-3 (1:500, Abcam, UK), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:1,000; Cell Signaling Technology, USA). The reactions were detected with a secondary horse-radish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG; 1:1,000; Abcam, UK).
Flow cytometry (FCM)
FCM using the Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit (Tianjin Sungene Biotech Co., Ltd.) was conducted to identify the apoptotic rate of LX-2 cells, in accordance with the manufacturer’s instructions.
Dual-luciferase experiment
Wild-type (WT) and mutant (mut) ARRB1-3' untranslated region (3'UTR) were cloned into the pGL3 vector to obtain the WT-pGL3-ARRB1-3'UTR and mut-pGL3-ARRB1-3'UTR vectors. The plasmids were transfected into human hepatic stellate LX-2 cells with miR-1295b-3p agomir or miR-NC (Gema, Shanghai, China) using Liposome 2000 (Invitrogen, Shanghai, China). After 48 h, the luciferase activity of the transfected cells was detected and analyzed.
Statistical analysis
Statistical analysis was performed with SPSS 22.0 software. Analysis of variance was used to compare the difference between groups. All experiments were repeated three times. A P value <0.05 was considered statistically significant.
Results
HE staining and Masson staining
HE staining
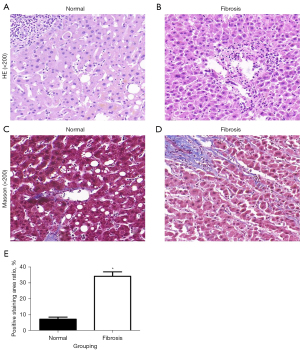
The anatomical structure of the normal liver was complete (Figure 1A). In contrast, the structure of the liver lobules and portal areas in liver fibrosis tissues were incomplete. In addition, the spindle cells were visible in immature collagen fibers (Figure 1B).
Masson staining
The collagen fibers in normal liver tissues were mainly distributed around the large blood vessels and portal system (Figure 1C). In contrast, the collagen fibers in the liver fibrosis tissue surrounded the blood vessels and portal veins and were increased in parts of the Disse space and between the liver lobules. In addition, the normal liver lobule structure was destroyed (Figure 1D) and the area of collagen in liver fibrosis tissue was significantly larger than that in normal liver tissues (P<0.05; Figure 1E).
Screening of differentially expressed microRNA
A total of 1,575 microRNAs were detected, of which 1,550 were known microRNAs, and 25 were newly discovered microRNAs. A total of 44 microRNAs were found to be differentially expressed, including 18 upregulated and 26 downregulated microRNAs, according to the conditions of P<0.05 and |log fold change (FC)| >1. Thus, cluster analysis (Figure 2) was performed to screen 5 downregulated microRNAs, including miR-483-3p, miR-5589-3p, miR-1271-5p, miR-1295b-3p, and miR-7977 (Table 2).
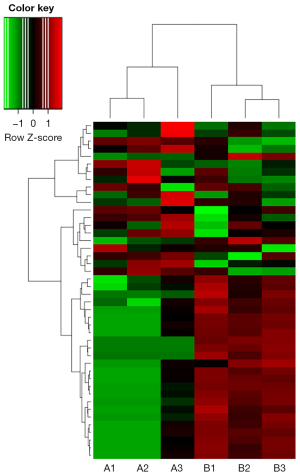
Table 2
| Gene | LogFC | LogCPM | P value | FDR |
|---|---|---|---|---|
| hsa-miR-483-3p | −6.511775741 | 5.032266762 | 0.0000056380 | 0.002982483 |
| hsa-miR-5589-3p | −7.685677736 | 2.073910688 | 1.0000056380 | 0.003135967 |
| hsa-miR-1271-5p | −5.492995496 | 2.398223531 | 2.0000056380 | 0.016749062 |
| hsa-miR-1295b-3p | −6.619455473 | 1.228638815 | 0.00024288 | 0.027765404 |
| hsa-miR-7977 | −6.92821172 | 1.464214675 | 0.000262433 | 0.027765404 |
CPM, counts-per-million; FC, fold change; FDR, false discovery rate.
Target gene prediction
The target genes of the differentially expressed microRNAs were predicted. The microRNAs miR-483-3p, miR-5589-3p, miR-1271-5p, miR-1295b-3p, and miR-7977 were all found to be downregulated in bile duct obstruction-induced liver fibrosis tissues, and had a negative regulatory effect on target genes. There were 127 target genes identified for miR-483-3p, 18 for miR-1271-5p, 15 for miR-5589-3p, 44 for miR-7977, and 14 for miR-1295b-3p (Table 3).
Table 3
| MicroRNA | Gene symbol |
|---|---|
| miR-483-3p | NOS1, NFASC, CPLX2, SSBP3, U2AF2, TAF5L, ARHGAP5, MLPH, ACSM4, RAP1GAP2, RNF24, DOLPP1, PARVG, GPT2, FKBP5, SMIM6, ISY1, ASB6, SLC24A1, NAPB, TET3, CREBL2, MTA2, TRIM62, PIK3C2B, HDAC3, CBX2, CCDC174, TMEM242, ADCY1, AUNIP, UCN2, LRRC46, COL11A2, NXNL1, ZNF699, TMEM179B, ACVR1C, EZR, MYRF, EXPH5, MAP7D3, RXFP1, TMEM128, RGL1, WSCD2, BNC2, ATAD1, ITGB8, GPATCH2L, PPP1R14C, SELENOI, CSF1, CDC42SE2, MYO5B, CBX5, SYK, PTPRQ, GFRA1, GPR17, DCTN5, WISP1, HSPA14, TRIB3, ZZZ3, DOK1, HCN4, ATG2B, TRIM56, NSMCE3, SPPL2C, OR5AR1, FAM168B, ABCF1, BCLAF1, CCDC17, C17orf53, CEACAM18, NTSR1, ARRB1, TCHH, KDM4D, KCTD15, SLX4, ITGB3, JAG1, ZIK1, ZYX, NSL1, TMEM120B, KNCN, BTBD9, TMEM192, TP73, SYNPO2, SPATC1, CISD3, SDK2, SLC44A2, TFDP2, APTX, PLRG1, NBL1, MINOS1-NBL1, MBD3, NDST1, MED20, TRAPPC2L, RNF220, SCAMP4, CBX7, SMARCC1, TAF15, HMGN2, HDAC4, IFI44L, LDOC1, SNX8, ZDHHC8, PALD1, APLN, TNFAIP1, GCC1, KDM7A, YIPF4, LGI3, GPAT4 |
| miR-1271-5p | CSF1, CDC42SE2, MYO5B, CBX5, SYK, PTPRQ, GFRA1, GPR17, DCTN5, WISP1, HSPA14, TRIB3, ZZZ3, DOK1, HCN4, ATG2B, TRIM56, NSMCE3 |
| miR-5589-3p | ACVR1C, EZR, MYRF, EXPH5, MAP7D3, RXFP1, TMEM128, RGL1, WSCD2, BNC2, ATAD1, ITGB8, GPATCH2L, PPP1R14C, SELENOI |
| miR-7977 | ITGB3, JAG1, ZIK1, ZYX, NSL1, TMEM120B, KNCN, BTBD9, TMEM192, EZR, TP73, SYNPO2, SPATC1, CISD3, SDK2, SLC44A2, TFDP2, APTX, PLRG1, NBL1, MINOS1-NBL1, MBD3, NDST1, MED20, TRAPPC2L, RNF220, SCAMP4, CBX7, SMARCC1, TAF15, HMGN2, HDAC4, IFI44L, LDOC1, SNX8, ZDHHC8, PALD1, APLN, TNFAIP1, GCC1, KDM7A, YIPF4, LGI3, GPAT4 |
| miR-1295b-3p | OR5AR1, FAM168B, ABCF1, BCLAF1, CCDC17, C17orf53, CEACAM18, NTSR1, ARRB1, TCHH, KDM4D, KCTD15, SLX4, SPPL2C |
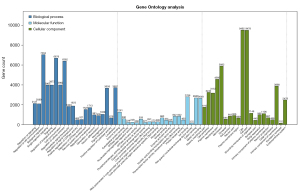
GO analysis
GO analysis of the target genes can be categorized into three aspects, its molecular function, location in the cell, and the biological processes in which it is involved (Figure 3). The target genes of the differentially expressed microRNAs were found to be mainly expressed on the plasma membrane. The proteins encoded by the target genes were associated with enzyme binding, ion (metal ion and cation) binding, transcription (specifically DNA binding and nucleic acid binding), cell localization, signal transduction, and biological regulation.
KEGG analysis
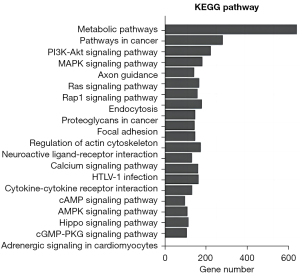
KEGG analysis was performed to determine the most important biochemical metabolic pathways and signal transduction pathways involving the candidate target genes. The top 20 most significant signal pathways were graphed (Figure 4). The target genes of the differentially expressed microRNAs primarily affected bile duct obstruction-induced liver fibrosis through metabolic pathways. These genes might also have a certain degree of influence on tumorigenesis.
miR-1295b-3p expression was downregulated in the bile duct obstruction-induced liver fibrosis
qRT-PCR was performed in the re-screened bile duct obstruction-induced liver fibrosis samples (n=5) and the normal liver samples (n=5). We found that miR-1295b-3p expression was downregulated in the fibrotic livers compared to the normal livers (Figure 5A).
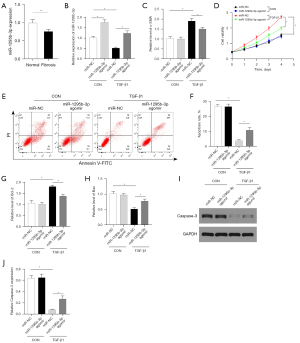
Overexpression of miR-1295b-3p can inhibit HSC activation and cell proliferation
Since miR-1295b-3p was downregulated in bile duct obstruction-induced liver fibrosis, this prompted us to further study the relationship between liver fibrosis and miR-1295b-3p. We used TGF-β1 to activate LX-2 cells, and transfected LX-2 cells with miR-NC or miR-1295b-3p agomir. qRT-PCR showed that the level of miR-1295b-3p in the TGF-β1-treated cells was lower than that in the control cells, while the level of miR-1295b-3p was upregulated after transfected with miR-1295b-3p agomir (Figure 5B). The expression of α-SMA was increased in the TGF-β1 group compared to the control group, while its expression was reduced after transfected with miR-1295b-3p agomir in the TGF-β1 group (Figure 5C). Moreover, CCK-8 assays demonstrated that the TGF-β1 treated group had an increased proliferative ability, but LX-2 cell viability was significantly reduced by miR-1295b-3p agomir (Figure 5D), suggesting that miR-1295b-3p inhibited LX-2 cell proliferation. Next, we studied whether miR-1295b-3p affected apoptosis of HSCs. FCM showed that compared with the miR-NC TGF-β1 group, miR-1295b-3p agomir increased the apoptosis of LX-2 cells (Figure 5E,5F). In addition, the mRNA levels of the pro-apoptotic protein Bax was increased, while the expression of the anti-apoptotic protein Bcl-2 was decreased after transfected with miR-1295b-3p agomir in TGF-β1 group (Figure 5G,5H). Moreover, Western blots showed that caspase-3 proteomic levels within the TGF-β1 group were obviously decreased, while miR-1295b-3p agomir upregulated caspase-3 protein expression (Figure 5I,5J). These results indicated that overexpression of miR-1295b-3p can inhibit HSCs activation and cell proliferation.
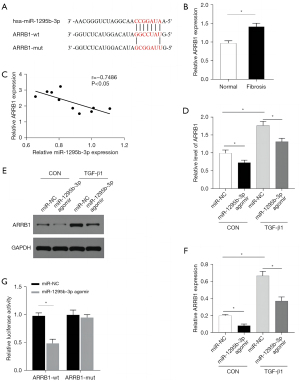
ARRB1 is a potential target of miR-1295b-3p
Bioinformatics analysis (TargetScan7.1) predicted the potential binding site of ARRB1 and miR-1295b-3p (Figure 6A). We analyzed the relationship between ARRB1 and miR-1295b-3p by detecting the expression of ARRB1 in human liver tissue. qRT-PCR analysis showed that ARRB1 expression was upregulated in liver fibrosis (Figure 6B). Moreover, Pearson’s correlation analysis showed that ARRB1 expression was inversely proportional to that of miR-1295b-3p (Figure 6C). As shown in Figure 6D, the expression of ARRB1 was increased in activated LX-2 cells, while overexpression of miR-1295b-3p decreased the level of ARRB1 in LX-2 cells. Moreover, Western blotting revealed that the protein level of ARRB1 was also upregulated in activated LX-2 cells, and overexpression of miR-1295b-3p downregulated the level of ARRB1 in LX-2 cells (Figure 6E,6F). The dual-luciferase assay was used to assess the relationship between ARRB1 and miR-1295b-3p. We transfected wild-type ARRB1 (ARRB1-wt) or mutant ARRB1 (ARRB1-mut) and miR-1295b-3p agomir or miR-NC into the LX-2 cells. The miR-1295b-3p agomir inhibited the luciferase activity of wild-type ARRB1 but had no significant effect on mutant ARRB1(Figure 6G). Therefore, miR-1295b-3p may regulate the expression of ARRB1 in bile duct obstruction-induced liver fibrosis.
Discussion
Bile duct obstruction-induced liver fibrosis is a liver disease caused by cholestasis, that results from an imbalance in tissue repair response (1). Bile duct obstruction causes cholestasis and increases bilirubin and bile acid levels in the serum. It leads to the inflammation of intrahepatic cells, promotes the activation of HSCs, and accelerates the activation of fibroblasts and the accumulation of inflammatory cells, ECM, and fibers in liver tissues. As a result, abnormal proliferation of connective tissues occurs. When the damage persists or worsens, the liver’s normal structure is destroyed under the action of related factors, eventually leading to changes in liver function and even failure (1,2). In recent years, the development of bioinformatics and epigenetics, as well as the discovery of microRNAs and long non-coding RNAs (lncRNAs) have gradually uncovered the role of TGF-β in bile duct obstruction-induced liver fibrosis (6-9). Kennedy et al. found that the level of miR-21 increased in rats with bile duct ligation; and inhibiting miR-21 expression could reduce HSC activation, collagen deposition, and the expression of TGF-β and α-SMA, thereby hindering the development of hepatic fibrosis (10).
In this current research, 5 differentially expressed microRNAs, miR-483-3p, miR-5589-3p, miR-1271-5p, miR-1295b-3p, and miR-7977, were found to be downregulated in bile duct obstruction-induced liver fibrosis. GO analysis and KEGG analysis further demonstrated that the target genes associated with the differentially expressed microRNAs were involved in metabolic and tumor pathways, such as the phosphoinositide 3-kinase (PI3K)-Akt, mitogen-activated protein kinase (MAPK), and Ras signaling pathways.
The down-regulation of miR-483-3p in bile duct obstruction-induced liver fibrosis leads to the up-regulation of BRCA1, causing cell cycle arrest and potentially affecting the formation of liver Mallory bodies and inhibiting liver cell regeneration (11,12). On the other hand, the overexpression of miR-483 acts on platelet-derived growth factor-B (PDGF-B) and tissue inhibitor of metalloproteinase 2 (TIMP2) during HSC activation, impeding carbon tetrachloride-induced liver fibrosis (13,14). The microRNA miR-1271-5p targets genes, such as zinc finger E-box-binding homeobox 1 (ZEB1), snail family transcriptional repressor 2 (SNAI2), phosphatases of regenerating liver 1 (PTP4A1), Forkhead box Q1 (FOXQ1), Forkhead box K2 (FOXK2), anaplastic lymphoma kinase (ALK) to inhibit epithelial-mesenchymal transition (EMT), thereby preventing the migration and invasion of hepatocellular carcinoma (HCC) cells (15). Yu et al. found that miR-1271-5p was upregulated in fibrotic human liver tissues and hindered autophagy of HSCs by reducing the expression of ATG7, thereby delaying the activation of HSCs (16). However, the role of miR-7977 and miR-5589-3p in liver diseases remain unclear (17-19). For the first time, this study demonstrated the differential expression of miR-7977 and miR-5589 in liver fibrosis induced by bile duct obstruction. We showed that miR-1295b-3p was downregulated in liver fibrosis. In vitro, miR-1295b-3p was downregulated in LX-2 cells induced by TGF-β1, and overexpression of miR-1295b-3p decreased the α-SMA levels and led to inhibition of proliferation and induction of apoptosis in LX-2 cells. The above research results suggested that overexpression of miR-1295b-3p alleviated liver fibrosis and inhibited HSCs activation.
ARRB1 is a gene in the β-arrestin family. β-arrestin is a scaffold protein that regulates G protein-coupled receptor (GPCR) signal transduction. It has a certain effect on cell proliferation via signal pathways such as Wnt, TGF-β, Notch, and autophagy, which in turn affects the deposition of the ECM, activates the inflammatory response, and the induces the development of fibrotic diseases (20,21). Xuan et al. found that overexpression of the genes encoding miR-29a or miR-652 decreased ARRB1 expression in mice with liver fibrosis and inhibited the development of liver fibrosis (22). ARRB1 overexpression promotes renal fibroblast activation and EMT through the typical Wnt1/β-catenin signaling pathway, accelerates renal fibrosis, affects the proliferation of hepatocytes through Akt signaling, and promotes the occurrence of liver cancer (23). Overexpression of β-arrestin in normal adult cardiac fibroblasts increases collagen synthesis through the ERK and Smad signaling pathways. In the early stages of myocardial infarction, ARRB1 expression is upregulated, activating myofibroblasts, increasing collagen synthesis and deposition, and exacerbating cardiac fibrosis (21). Li et al. verified in animal experiments that melatonin (MT) enhances protection of non-alcoholic fatty liver disease by activating ARRB1 (24). In this study, ARRB1 was predicted to be the target gene of miR-1295b-3p, which may regulate the occurrence and development of bile duct obstruction-induced liver fibrosis. The ARRB1 expression levels in human fibrotic liver specimens and activated LX-2 cells were all upregulated. Furthermore, overexpression of miR-1295b-3p in vitro inhibited ARRB1 expression. Luciferase experiments showed that miR-1295b-3p regulates the expression of ARRB1 by directly binding to the 3'UTR of its mRNA. However, the specific mechanisms and functions remain unclear, and warrants further investigation.
Conclusions
In summary, the changes in the expression level of microRNA in bile duct obstruction-induced liver fibrosis and its relationship with and the mechanisms of bile duct obstruction-induced liver fibrosis are still unclear. However, many studies have shown that microRNAs can promote or inhibit inflammation, as well as the activation, proliferation, and apoptosis of HSCs, and the accumulation of the ECM. This study, based on gene sequencing, compared normal and bile duct obstruction-induced liver fibrosis tissues to obtain the differentially expressed microRNAs and the target genes. Bioinformatics was used to study its corresponding functions and pathways in the process of bile duct obstruction-induced liver fibrosis. Moreover, further studies showed the inhibitory effect of miR-1295b-3p on liver fibrosis, and we discovered that ARRB1 may be a potential target of miR-1295b-3p. However, the regulatory mechanisms require further investigation. The differentially expressed microRNAs may be novel biological markers for the diagnosis, treatment, and prognosis of bile duct obstruction-induced liver fibrosis.
Acknowledgments
The authors wish to thank all colleagues, the reviewers, and the editors for improving our paper.
Funding: This work was supported by the China Postdoctoral Science Foundation (project No. 2018M633732).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6416/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6416/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6416/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Qingdao Municipal Hospital (2020 No. 098) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gulamhusein AF, Hirschfield GM. Primary biliary cholangitis: pathogenesis and therapeutic opportunities. Nat Rev Gastroenterol Hepatol 2020;17:93-110. [Crossref] [PubMed]
- Zhang M, Serna-Salas S, Damba T, et al. Hepatic stellate cell senescence in liver fibrosis: Characteristics, mechanisms and perspectives. Mech Ageing Dev 2021;199:111572. [Crossref] [PubMed]
- Villesen IF, Daniels SJ, Leeming DJ, et al. Review article: the signalling and functional role of the extracellular matrix in the development of liver fibrosis. Aliment Pharmacol Ther 2020;52:85-97. [Crossref] [PubMed]
- Huang YH, Kuo HC, Yang YL, et al. MicroRNA-29a is a key regulon that regulates BRD4 and mitigates liver fibrosis in mice by inhibiting hepatic stellate cell activation. Int J Med Sci 2019;16:212-20. [Crossref] [PubMed]
- Yang X, Ma L, Wei R, et al. Twist1-induced miR-199a-3p promotes liver fibrosis by suppressing caveolin-2 and activating TGF-β pathway. Signal Transduct Target Ther 2020;5:75. [Crossref] [PubMed]
- Dewidar B, Meyer C, Dooley S, et al. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells 2019;8:1419. [Crossref] [PubMed]
- Gupta P, Sata TN, Yadav AK, et al. TGF-β induces liver fibrosis via miRNA-181a-mediated down regulation of augmenter of liver regeneration in hepatic stellate cells. PLoS One 2019;14:e0214534. [Crossref] [PubMed]
- Liu W, Feng R, Li X, et al. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging (Albany NY) 2019;11:9569-80. [Crossref] [PubMed]
- Yang Y, Sun M, Li W, et al. Rebalancing TGF-β/Smad7 signaling via Compound kushen injection in hepatic stellate cells protects against liver fibrosis and hepatocarcinogenesis. Clin Transl Med 2021;11:e410. [Crossref] [PubMed]
- Kennedy LL, Meng F, Venter JK, et al. Knockout of microRNA-21 reduces biliary hyperplasia and liver fibrosis in cholestatic bile duct ligated mice. Lab Invest 2016;96:1256-67. [Crossref] [PubMed]
- Zhang K, Fu W, Zhao S, et al. miR-483-3p Promotes IL-33 Production from Fibroblast-Like Synoviocytes by Regulating ERK Signaling in Rheumatoid Arthritis. Inflammation 2021;44:2302-8. [Crossref] [PubMed]
- Liu H, French BA, Li J, et al. Altered regulation of miR-34a and miR-483-3p in alcoholic hepatitis and DDC fed mice. Exp Mol Pathol 2015;99:552-7. [Crossref] [PubMed]
- Liu D, Liu F, Li Z, et al. HNRNPA1-mediated exosomal sorting of miR-483-5p out of renal tubular epithelial cells promotes the progression of diabetic nephropathy-induced renal interstitial fibrosis. Cell Death Dis 2021;12:255. [Crossref] [PubMed]
- Huang G, Zhang J, Qing G, et al. Downregulation of miR-483-5p inhibits TGF-β1-induced EMT by targeting RhoGDI1 in pulmonary fibrosis. Mol Med Rep 2021;24:538. [Crossref] [PubMed]
- Liu HM, Tan HY, Lin Y, et al. MicroRNA-1271-5p inhibits cell proliferation and enhances radiosensitivity by targeting CDK1 in hepatocellular carcinoma. J Biochem 2020;167:513-24. [Crossref] [PubMed]
- Yu K, Li N, Cheng Q, et al. miR-96-5p prevents hepatic stellate cell activation by inhibiting autophagy via ATG7. J Mol Med (Berl) 2018;96:65-74. [Crossref] [PubMed]
- Yoshida M, Horiguchi H, Kikuchi S, et al. miR-7977 inhibits the Hippo-YAP signaling pathway in bone marrow mesenchymal stromal cells. PLoS One 2019;14:e0213220. [Crossref] [PubMed]
- Chen L, Cao P, Huang C, et al. Serum exosomal miR-7977 as a novel biomarker for lung adenocarcinoma. J Cell Biochem 2020;121:3382-91. [Crossref] [PubMed]
- Zhang X, Zhang G, Huang H, et al. Differentially Expressed MicroRNAs in Radioresistant and Radiosensitive Atypical Meningioma: A Clinical Study in Chinese Patients. Front Oncol 2020;10:501. [Crossref] [PubMed]
- Tan S, Lu Y, Xu M, et al. β-Arrestin1 enhances liver fibrosis through autophagy-mediated Snail signaling. FASEB J 2019;33:2000-16. [Crossref] [PubMed]
- Philip JL, Xu X, Han M, et al. Regulation of cardiac fibroblast-mediated maladaptive ventricular remodeling by β-arrestins. PLoS One 2019;14:e0219011. [Crossref] [PubMed]
- Xuan J, Guo SL, Huang A, et al. MiR-29a and miR-652 Attenuate Liver Fibrosis by Inhibiting the Differentiation of CD4+ T Cells. Cell Struct Funct 2017;42:95-103. [Crossref] [PubMed]
- Xu C, Xu Z, Zhang Y, et al. β-Catenin signaling in hepatocellular carcinoma. J Clin Invest 2022;132:e154515. [Crossref] [PubMed]
- Li DJ, Tong J, Li YH, et al. Melatonin safeguards against fatty liver by antagonizing TRAFs-mediated ASK1 deubiquitination and stabilization in a β-arrestin-1 dependent manner. J Pineal Res 2019;67:e12611. [Crossref] [PubMed]
(English Language Editor: J. Teoh)