
The m6A demethylase FTO promotes keloid formation by up-regulating COL1A1
Highlight box
Key findings
• FTO regulates the expression of COL1A1 by down regulating m6a level in skin fibroblasts.
What is known and what is new?
• m6A modification has gradually been proven to be related to keloid and can be used as a potential target for scar therapy;
• The effect and mechanism of m6A modified enzymes in keloid are still need to be further studied.
What is the implication, and what should change now?
• FTO and dynamic regulation of m6A are involved in the development of keloid, and can be used as the target of keloid.
Introduction
Keloid is a dermal fibrotic disease characterized by excessive proliferation of dermal fibroblasts and deposition of collagen (1). Keloid usually occurs in areas of the body where the skin is constantly tensed and stretched, such as the anterior chest and scapular region, and less frequently on the scalp and anterior calf (2). In clinical practice, keloid can cause both physical and psychological damage to the patient (3). Keloid can be generically categorized into 2 groups, primary and secondary, but the etiology is unknown. Radiation therapy, compression therapy, cryosurgery, and pharmaceutical treatments such as glucocorticoids are among the available treatment options (4,5). However, these treatments are not particularly effective. Therefore, it is vitally crucial to explore new potential pathogeneses and develop novel treatment strategies.
N6-methyladenosine (m6A) modification, the most abundant RNA modification, belongs to an important mechanism of post-transcriptional regulation that plays a critical role in a wide range of physiological and pathological processes, including obesity, cardiovascular disease, azoospermia or oligospermia, intervertebral disc degeneration, and cancer metabolism (6-8). m6A modification regulate mRNA splice, stability and translation. The critical regulatory role of m6A modifications in keloid tissue formation has also garnered growing attention from researchers (9,10). The present research focuses on demethylase, and finds that Fat mass and obesity-associated protein (FTO) promotes keloid formation. FTO is one of the most significant demethylating enzymes, the function of which is to demethylate bases that have previously been changed by m6A, hence regulating bodily activities (11-13). Nonetheless, the significance of FTO in the regulation of keloid development remains unclear. Therefore, this study aimed to investigate the specific regulatory role of FTO in the process of keloid development, with the hope of identifying new therapeutic targets for keloids in the future. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6021/rc).
Methods
Tissues and skin fibroblasts isolation
This study was approved by the Medical and Ethics Committees of the First Affiliated Hospital of Shantou University Medical College (No. B-2022-055), and each patient signed an informed consent before enrolling in the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A total of 15 patients with keloid were recruited from 2021 to 2022 at the Plastic and Aesthetic Surgery Center in the First Affiliated Hospital of Shantou University Medical College, and the normal group samples were obtained from the adjacent to the lesion in these 15 keloid cases. Normal skin tissue was separated into 1 mm3 pieces. The pieces were then digested with 0.25% trypsin (#25200072, Thermo Fisher Scientific, Waltham, MA, USA) for 3 minutes. After washing and mechanical separation, the fibroblasts were filtered, centrifuged (600 ×g for 6 minutes), and resuspended in Dulbecco’s modified Eagle medium (DMEM; #10313021, Thermo Fisher Scientific) with 10% fetal bovine serum (FBS) in a cell culture incubator (37 ℃, 5% CO2). The medium was changed every 2–3 days. Experiments subsequently utilized fibroblasts from generations 3 to 6.
Antibodies and reagents
Antibody against m6A (#SAB5600251) was acquired from Merck (Burlington, MA, USA). Antibodies against FTO (#27226-1-AP), ALKB homolog 5 (ALKBH5) (#16837-1-AP), α-smooth muscle actin (α-SMA) (#14395-1-AP), collagen type I alpha 1 chain (COL1A1) (#67288-1-Ig), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#60004-1-Ig) were purchased from Proteintech (Rosemont, IL, USA).
Hematoxylin and eosin (H&E) staining
Paraffin-embedded skin tissues were cut into sections and placed on slides. The slides were degreased with xylene and then dehydrated with different dilutions of ethanol (100%, 95%, 85%, 70%, 50%). The slides were stained with hematoxylin (#BL735A-1, Biosharp, Beijing, China) for 5 minutes, rinsed with water for 2 minutes, and then treated with ethanol for 10 seconds. The slides were then stained with eosin (#BL735A-2, Biosharp) and incubated for 10–15 seconds. An Olympus BX63 microscope (Olympus, Tokyo, Japan) was used to acquire images of the stained slides.
m6A dot blotting
Total RNA was purified from skin tissues and diluted to different concentrations. RNA was denatured at 95 ℃ and then incubated on ice to inhibit the creation of the messenger RNA’s (mRNA’s) secondary structure. RNA was transferred onto the fiber membrane (#ISEQ00010, Merck Millipore, Burlington, MA, USA). The membrane was subsequently exposed to ultraviolet (UV) light. The membrane was rinsed for 5 minutes with tris-buffered saline with Tween 20 (TBST; Biosharp) and then incubated with an anti-m6A antibody (Millipore, Burlington, MA, USA) at 4 ℃. The membrane was then rinsed 3 times for 5 minutes with TBST and incubated with goat anti-rabbit IgG-horseradish peroxidase (HRP) antibody (#BL003A, Biosharp) at room temperature. After that, the membrane was washed 3 times (10 minutes each time) with TBST and incubated with Western blotting substrate (Biosharp) at room temperature. Finally, the dots were viewed using a microscope (OI-X6, Olympus).
Real-time RT-PCR assay
TRIzol (Solarbio, Beijing, China) was used to extract total RNA from cultured fibroblasts. The RNA concentrations were measured using the NanoDrop 2000 instrument (Bio-Rad, Hercules, CA, USA). The complementary DNAs (cDNAs) were subsequently synthesized from 1 µg total RNA using an RNA Reverse Transcription Kit (APExBIO, Houston, TX, USA). Then, a quantitative reverse transcription polymerase chain reaction (qRT-PCR) experiment was performed using the LightCycler 480 SYBR Green I Master Kit (APExBIO). The mRNA expression levels were determined using the standard 2−ΔΔCt method based on at least 3 biological replicates. The primer sequences of different genes are shown in Table 1.
Table 1
| Gene | Primer sequence (5'-3') |
|---|---|
| GAPDH | Forward: GAGTCAACGGATTTGGTCGT |
| Reverse: GACAAGCTTCCCGTTCTCAG | |
| ALKBH5 | Forward: CGCCGTCATCAACGACTACC |
| Reverse: CGACACCCGAATAGGCTTG | |
| FTO | Forward: TTGCCCGAACATTACCTGCT |
| Reverse: TGTGAGGTCAAACGGCAGAG | |
| α-SMA | Forward: ACTGCCTTGGTGTGTGACAA |
| Reverse: CACCATCACCCCCTGATGTC | |
| COL1A1 | Forward: AGCAGACTGGCAACCTCAAGA |
| Reverse: GGGAGGTCTTGGTGGTTTTG | |
| Position 1 | Forward: AGATGGCATCCCTGGACAG |
| Reverse: CGGTTGATTTCTCATCATAGCC | |
| Position 2 | Forward: CTGGCTCTCCTGGTGAACA |
| Reverse: ACCGTTGAGTCCATCTTTGC | |
| Position 3 | Forward: CAACCTGGATGCCATCAA |
| Reverse: ACATGCCTCTTGTCCTTG | |
| Position 4 | Forward: ACAGACAAGCAACCCAAA |
| Reverse: TGAGAGATGAATGCAAAGGA | |
| Position 5 | Forward: TCGGAGGAGAGTCAGGAA |
| Reverse: CAGCAACACAGTTACACAAG |
qPCR, quantitative polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ALKBH5, ALKB homolog 5; COL1A1, collagen type I alpha 1 chain; α-SMA, α-smooth muscle actin; FTO, fat mass and obesity-associated protein.
Cell transfection
Fibroblasts were cultured in a 6-well plate. The coding sequence (CDS) genes for FTO and ALKHB5 were synthesized and inserted into pCDH overexpression vectors (designed and constructed by Sangon Biotech, Shanghai, China). Fibroblasts were transfected with the relevant plasmids using Lipofectamine 2000 (#BL623B, Biosharp) according to the manufacturer’s instructions. After 24 hours of transfection, the effectiveness of overexpression was determined by qPCR and Western blotting.
Transwell migration assay
After 24 hours of fibroblast starvation, the cells were digested and the cell density was adjusted to 2×105 cells per milliliter. After adding 100 µL of cell suspension to the transwell chamber, 500 µL of DMEM containing 20% FBS was added to the lower chamber. After 48 hours of cell culture, non-migrating cells were removed from the upper chamber with wet cotton balls, and the migrating cells were stained with crystal violet (#BS941-5g, Biosharp) for 20 minutes and counted.
Western blotting assay
Total protein samples were extracted from cultured fibroblasts using radioimmunoprecipitation assay (RIPA) solution (Biosharp). Protein concentrations were determined using a bicinchoninic acid (BCA) kit (#BL521A, Biosharp), and protein samples were separated using 6.5–12.5% polyacrylamide gel electrophoresis (PAGE) Gel Fast Preparation Kits (#BL566B, Biosharp) and transferred onto polyvinylidene fluoride (PVDF) membranes (#ISEQ00010, Merck Millipore). The membranes were blocked with 5% non-fat milk (#A600669, Sangon Biotech) for 1 hour, then incubated with anti-FTO (1:1,000, Proteintech), anti-GAPDH (1:2,000, Proteintech), anti-COL1A1 (1:1,000, Proteintech), and anti-α-SMA (1:1,000, Proteintech) overnight at 4 ℃. The membranes were then incubated with goat anti-rabbit or anti-mouse IgG-HRP antibody (1:2,000, Proteintech) for 1 hour. Blots were visualized using a commercial enhanced chemiluminescent reagent (#BL523B-1, Biosharp).
MeRIP-qPCR
Total RNA was extracted using TRIzol and then using a methylated RNA immunoprecipitation kit (Merck Millipore) according to the manufacturer’s instructions. Briefly, 25 µg of mRNA was broken into 100 nt RNA fragments by heating, and then incubated with a specifically recognized m6A antibody for 2 hours. The antibody-bound RNA fragments were purified by adding protein A/G magnetic beads, the immunoprecipitated RNA was reverse-transcribed into cDNA, and an RT-PCR amplification reaction with a cDNA template was performed. The amplified PCR products were subjected to lysis curve analysis, and the data were subsequently analyzed using ABI Prism 7300 SDS software (Thermo Fisher Scientific).
Statistical analysis
Data from various groups were analyzed using independent t-tests. Experiments were repeated in triplicate. The GraphPad Prism 9.0 software (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Statistical significance was indicated when P<0.05: “*” represents P<0.05; “**” represents P<0.01; “***” represents P<0.001.
Results
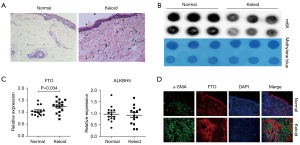
m6A modification levels were down-regulated, whereas m6A demethylase FTO expression was up-regulated in keloid tissues
In this study, we found that the dermis of keloid patients’ skin was significantly hyperplastic and hypertrophic with disturbed cell organization by comparing H&E staining of keloid patients’ skin tissues with normal human skin tissues (Figure 1A). We then assessed the level of RNA m6A modification in keloid tissues and normal tissues by m6A dot blotting assay and discovered that it was dramatically decreased in keloid tissues (Figure 1B). Since FTO and ALKBH5 are the 2 most important m6A demethylases, we examined the mRNA expression of these 2 molecules in keloid tissues (N=15) and normal tissues (N=15). We found that the expression of FTO increased in keloid tissues, whereas the expression of ALKBH5 did not change significantly (Figure 1C), indicating that FTO may play a regulatory role in the keloid formation process. Using the FTO and α-SMA co-localization test, the expression of demethylase FTO was dramatically up-regulated (Figure 1D). These findings show that m6A modification levels are decreased in keloid tissue, but m6A demethylase FTO expression is increased.
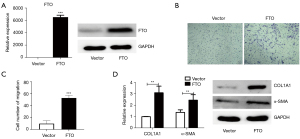
m6A demethylase FTO stimulates cell migration and collagen deposition of skin fibroblasts
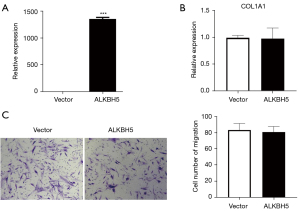
To further validate the function of FTO, we overexpressed the FTO molecule in skin fibroblasts (Figure 2A), and discovered that this dramatically increased fibroblast migration (Figure 2B,2C). Considering the importance of excessive collagen deposition and tissue fibrosis in keloid tissue formation, we next detected the mRNA and protein levels of the collagen deposition marker COL1A1 and the fibrosis marker α-SMA and found that overexpression of FTO significantly promoted these 2 molecules (Figure 2D), suggesting that FTO may promote the formation of keloid by upregulating the expression of COL1A1. In addition, we discovered that overexpression of ALKBH5, another major m6A demethylase, had no influence on the migration and collagen deposition of skin fibroblasts (Figure 3), indicating that this molecule may not be a key regulator of this process. The above results indicate that the m6A demethylase FTO enhances the cell migration of dermal fibroblasts and the deposition of collagen in order to contribute to the development of keloids.
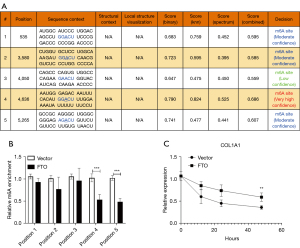
FTO regulates COL1A1 m6A modification and maintains mRNA stability
To confirm how FTO regulates the COL1A1 molecule and mediates keloid formation, we utilized the SRAMP database (http://www.cuilab.cn/sramp/) to predict that COL1A1 contains 5 essential m6A modification positions (Figure 4A). Then, after overexpression of FTO, we detected 5 m6A modification positions and found that the expression levels of m6A modification position 4 and position 5 of COL1A1 decreased (Figure 4B). Further, using the actinomycin D to study the stability of COL1A1 mRNA, we found that overexpression of FTO inhibited the degradation rate of COL1A1 mRNA, hence maintaining mRNA stability (Figure 4C). In conclusion, FTO regulates COL1A1 m6A modification and maintains the stability of COL1A1 mRNA to contribute to the regulation of keloid formation.
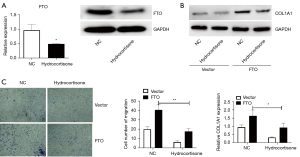
Glucocorticoids can improve keloids by suppressing the expression of FTO
In the clinic, glucocorticoids are among the most commonly used drugs for the treatment of keloid. However, it is unknown whether FTO mediates their therapeutic effects. After 48 hours of treatment with glucocorticoids on skin fibroblasts, we showed that glucocorticoids can significantly suppress the expression of FTO (Figure 5A). After overexpression of FTO, glucocorticoid treatment could inhibit the up-regulation of COL1A1 mRNA expression induced by FTO overexpression (Figure 5B). In the further transwell experiment, it was discovered that overexpression of FTO followed by glucocorticoid treatment significantly suppressed cell migration of skin fibroblasts (Figure 5C). The above findings demonstrate that glucocorticoids can improve keloids by reducing the expression of FTO.
Discussion
In this study, we observed a drop in m6A modification levels and an increase in m6A demethylase FTO expression in keloid tissue. Subsequently, overexpression of FTO stimulated the migration of skin fibroblasts and the expression of COL1A1 and α-SMA. Mechanistic experiments confirmed that FTO promotes keloid development by modulating m6A modification and mRNA stability of COL1A1. This study also demonstrated that FTO is involved in the treatment of keloids with glucocorticoids. In conclusion, our study demonstrates that FTO upregulates the expression of COL1A1 by regulating the m6A modification of COL1A1 and maintaining mRNA stability, hence promoting the formation of keloids, which may provide a novel target for the future treatment of keloids.
FTO is one of the most significant demethylases, which not only regulates many physiological processes but also plays a critical regulatory function in the formation of keloids. In this study, we discovered that the expression of FTO was up-regulated in keloid tissue, and that overexpression of FTO may increase the migration of fibroblasts and the expression of COL1A1 and α-SMA. This fully demonstrates that FTO regulates keloid development by promoting collagen deposition and fibroblast migration. As far as we know, this is the first study to explore the regulatory relationship between FTO and keloids, providing a theoretical experimental foundation for future keloid treatment research.
COL1A1 also plays a crucial regulatory role in the formation of keloids. In this study, the expression changes of the m6A modification site of COL1A1 and the stability of COL1A1 mRNA were shown to mediate the regulatory effect of FTO on keloids. Previous study has demonstrated that COL1A1 can promote the formation of keloids since excessive collagen deposition is an essential characteristic of keloids. A Linjawi et al. discovered that the COL1A1 −1,997 G/T (rs1107946) and Sp1 +1,245 G/T (rs1800012) gene promoters showed a strong positive connection with the risk of keloid formation (14). Wang et al. found that COL1A1 mediates the promotion of H19 on the proliferation and metastasis of skin fibroblasts (15). In addition, Jung et al. discovered that COL1A1 is important in the regulation of sphingosine-1-phosphate (S1P) on keloid formation (16). In conclusion, COL1A1 contributes to the process of keloid formation. Signal pathway, like transforming growth factor β (TGF-β) and Wnt, may be modified and regulated by m6A to participate in keloid formation.
Glucocorticoids are among the most commonly used drugs for treating keloids. Our study revealed that FTO mediates the therapeutic effect of glucocorticoids on keloids. This study indicated that glucocorticoid treatment significantly inhibited the expression of FTO. In addition, glucocorticoids suppressed the up-regulation of COL1A1 and inhibited cell migration induced by FTO overexpression. This demonstrates that glucocorticoids can treat keloids by decreasing the expression of FTO. Additionally, previous study has demonstrated that FTO mediates the crucial regulation of glucocorticoid function. According to Hu et al., glucocorticoid-mediated FTO transactivation induces lipid accumulation in hepatocytes (17). In addition, Engel et al. discovered that the regulation of N-methyladenosine (mA) and N,2'-O-dimethyladenosine (mAm) was impaired in major depression patients following glucocorticoid stimulation, which also indicates a dysfunction of the demethylase FTO (18). The above findings indicate that FTO not only mediates the therapeutic effect of glucocorticoids on keloids, but also mediates the effects of glucocorticoids on other bodily functions.
Our study had several limitations. First, the molecular mechanism of this study was relatively superficial, and the alterations in downstream pathway molecules resulting from the up-regulation of COL1A1 have not been fully elucidated. In addition, this study lacked animal experiments, which will be the direction our research group takes to further investigate in the future.
Conclusions
In this study, we observed a drop in m6A modification levels and an increase in m6A demethylase FTO expression in keloid tissue. Subsequently, overexpression of FTO stimulated the migration of skin fibroblasts and the expression of COL1A1 and α-SMA. Mechanistic experiments confirmed that FTO promotes keloid formation by modulating m6A modification and mRNA stability of COL1A1. This study also demonstrated that FTO is involved in the treatment of keloids with glucocorticoids. In conclusion, our study demonstrates that FTO increases the expression of COL1A1 by regulating the m6A modification of COL1A1 and maintaining mRNA stability, hence increasing the formation of keloids.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6021/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6021/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6021/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Limandjaja GC, Niessen FB, Scheper RJ, et al. The Keloid Disorder: Heterogeneity, Histopathology, Mechanisms and Models. Front Cell Dev Biol 2020;8:360. [Crossref] [PubMed]
- Ogawa R, Okai K, Tokumura F, et al. The relationship between skin stretching/contraction and pathologic scarring: the important role of mechanical forces in keloid generation. Wound Repair Regen 2012;20:149-57. [Crossref] [PubMed]
- Shan M, Wang Y. Viewing keloids within the immune microenvironment. Am J Transl Res 2022;14:718-27. [PubMed]
- Liu XG, Zhang D. Evaluation of Efficacy of Corticosteroid and Corticosteroid Combined with Botulinum Toxin Type A in the Treatment of Keloid and Hypertrophic Scars: A Meta-Analysis. Aesthetic Plast Surg 2021;45:3037-44. [Crossref] [PubMed]
- Kumar AS, Kamalasanan K. Drug delivery to optimize angiogenesis imbalance in keloid: A review. J Control Release 2021;329:1066-76. [Crossref] [PubMed]
- Kumari R, Ranjan P, Suleiman ZG, et al. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res 2022;118:1680-92. [Crossref] [PubMed]
- Chen X, Wang J, Tahir M, et al. Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases. Cell Biosci 2021;11:147. [Crossref] [PubMed]
- An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer 2022;21:14. [Crossref] [PubMed]
- Lv W, Ren Y, Hou K, et al. Epigenetic modification mechanisms involved in keloid: current status and prospect. Clin Epigenetics 2020;12:183. [Crossref] [PubMed]
- Xie J, Zhang X, Zhang K, et al. Construction and validation of the diagnostic model of keloid based on weighted gene co-expression network analysis (WGCNA) and differential expression analysis. J Plast Surg Hand Surg 2022; Epub ahead of print. [Crossref] [PubMed]
- Li Y, Su R, Deng X, et al. FTO in cancer: functions, molecular mechanisms, and therapeutic implications. Trends Cancer 2022;8:598-614. [Crossref] [PubMed]
- Xie G, Wu XN, Ling Y, et al. A novel inhibitor of N (6)-methyladenosine demethylase FTO induces mRNA methylation and shows anti-cancer activities. Acta Pharm Sin B 2022;12:853-66. [Crossref] [PubMed]
- Relier S, Rivals E, David A. The multifaceted functions of the Fat mass and Obesity-associated protein (FTO) in normal and cancer cells. RNA Biol 2022;19:132-42. [Crossref] [PubMed]
- A Linjawi S, E Tork S, M Shaibah R. Genetic association of the COL1A1 gene promoter -1997 G/T (rs1107946) andSp1 +1245 G/T (rs1800012) polymorphisms and keloid scars in a Jeddah population. Turk J Med Sci 2016;46:414-23. [PubMed]
- Wang Z, Feng C, Song K, et al. lncRNA-H19/miR-29a axis affected the viability and apoptosis of keloid fibroblasts through acting upon COL1A1 signaling. J Cell Biochem 2020;121:4364-76. [Crossref] [PubMed]
- Jung SH, Song YK, Chung H, et al. Association between sphingosine-1-phosphate-induced signal transduction via mitogen-activated protein kinase pathways and keloid formation. Arch Dermatol Res 2019;311:711-9. [Crossref] [PubMed]
- Hu Y, Feng Y, Zhang L, et al. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m(6)A on lipogenic mRNAs. RNA Biol 2020;17:930-42. [Crossref] [PubMed]
- Engel M, Eggert C, Kaplick PM, et al. The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 2018;99:389-403.e9. [Crossref] [PubMed]
(English Language Editor: J. Jones)


