
Construction of a TFs-miRNA-mRNA network related to idiopathic pulmonary fibrosis
Highlight box
Key findings
• We construct a TFs-miRNA-mRNA network related to IPF and verify TFs by qRT-PCR.
What is known and what is new?
• Many transcription factors (TFs), microRNAs and mRNAs play important roles in IPF.
• We construct a TFs-miRNA-mRNA network related to IPF and perform functional enrichment analysis.
What is the implication, and what should change now?
• The novel TFs-miRNA-mRNA network provide new insights into the underlying molecular mechanisms of IPF.
• ARNTL, TRIM28, EZH2, BCOR, ASXL1, BCL6, BHLHE40, FOXA1, and EGR1 may become effective biomarkers for diagnosis and treatment of IPF.
Introduction
Idiopathic pulmonary fibrosis (IPF), the most common idiopathic interstitial pneumonia, is a chronic progressive fibrotic lung disease of unknown etiology that occurs predominantly in elderly adults, with a median age at diagnosis of 65 years old (1). It is associated with a high mortality rate, and therapies that slow disease progression are now available (2). Overall, patients with IPF have a similar life expectancy to those with non-small cell lung cancer, with a reported median survival rate estimate of 50% at 3 years and 20% at 5 years post-diagnosis (3,4). In recent years, the prevalence of IPF and the rates of hospital admissions and deaths due to the disease appear to be increasing, suggesting an increasing disease burden (5-7). Therefore, there is an urgent need to further understand the pathogenesis of IPF and identify novel markers to provide new research ideas for clinical treatment.
Historically, IPF was considered a chronic inflammatory disease, which gradually progresses to fibrosis. However, this concept was reassessed following the recognition that anti-inflammatory treatment did not improve disease outcomes, and the immunosuppressive treatment strategy of prednisolone and azathioprine was shown to increase mortality (8,9). Currently, IPF is generally considered to be the result of the interaction of multiple genetic and environmental risk factors that repeatedly act on the local alveolar epithelium to cause micro-injury. These micro-injuries trigger aberrant epithelial-fibroblast communication, induce myofibroblast activation, and cause massive extracellular matrix deposition and interstitial remodeling (10).
In recent years, microRNA (miRNA) has gained increasing attention from researchers in the field of IPF. Several reports have revealed the critical roles of miRNA and targeted messenger RNA (mRNA) in the development of various diseases, including IPF. Moreover, the expression of target genes could also be regulated by transcription factors (TFs) combining cis-elements at promoter locations (11). It is known that some TFs are involved in the pathogenesis of IPF. For example, early growth response transcription factors were found to be key mediators of fibrosis (12). Moreover, epithelial-mesenchymal transition (EMT) is known as an important feature in IPF and it is regulated by several TFs, which are members of prominent families of master regulators of transcription, such as the Forkhead family (FOXO), Twist family (TWIST), SNAIL family of zinc-finger transcription factors (SNAIL) and zinc-finger E-box-binding family (ZEB) (13). Thus, TFs may play important roles in the development of IPF. Many studies showed that transforming growth factor‑β1 (TGFβ1) plays a central role in the development of IPF (14). TGFβ1 promotes the fibrotic process of IPF through a variety of signaling pathways, including the MAPK, Smad and ERK signaling pathways (15-18). For example, transforming growth factor (TGF)-β induces SMAD family member 3 (SMAD3), which inhibits let-7d expression by binding to the upstream region of let-7 (19). The down-regulated expression of let-7 and consequent over-expression of high mobility group AT-hook 2 (HMGA2) decreases the expression of epithelial markers, cytokeratin and tight junction protein 1 (TJP1), and increases the expression of mesenchymal markers, actin alpha 2 (ACTA2) and vimentin (VIM), thereby inducing EMT to promote pulmonary fibrosis (20,21). Liu et al. demonstrated that TGF-β signaling leads to significant overexpression of microRNA-21 (miR-21) in the lungs of bleomycin-induced mice, which acts as an amplifying circuit to increase the fibrotic activity of TGF-β (22). Yang et al. showed that miR-200 family members (200a, 200b, 200c) inhibit EMT and reverse the fibrotic function of lung fibroblasts (23). Yang et al. also showed that miR-200 family members act as negative regulators of TGF-β-mediated pulmonary fibrosis, attenuate the expression of TGF-β-mediated mesenchymal markers, and can be candidates for the treatment of pulmonary fibrosis (23). Zhu et al. constructed a miRNA-mRNA network in IPF (24), but the reason for differential mRNAs and miRNAs was not explored. We conjectured that with so many differentially expressed mRNAs and miRNAs in IPF, it is likely that broad-spectrum regulatory factors such as TFs are involved upstream. Therefore, we predicted the possible regulatory causes of mRNA and miRNA differential expression by constructing a TFs-miRNA-mRNA regulatory network, thus facilitating further research exploration in the future. In addition, in the current study, the approach of using WGCNA can better focus on the most important gene modules within IPF tissue samples and provide more accurate and reliable downstream genes to build the network compared with simple differential gene intersection. Therefore, constructing the TFs-miRNA-mRNA regulatory network in IPF is really important and necessary.
In this study, we first screened several differentially expressed miRNAs (DEMs) in the lung tissues of IPF patients and compared these with normal tissues by analyzing the GSE13316 dataset downloaded from the Gene Expression Omnibus (GEO) database. Then, DE-mRNAs [differentially expressed genes (DEGs)] between IPF tissues and normal tissues were obtained using the GSE110147 and GSE53845 datasets downloaded from the GEO database. Gene set enrichment analysis (GSEA) of the DEGs was conducted using the clusterprofiler R package (25). To screen an important module in DEGs, weighted gene co-expression network analysis (WGCNA) analysis of the DEGs was performed using the “WGCNA” R package (26). Subsequently, Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (27,28) of the important module genes were conducted using the clusterprofiler R package. Next, the miRNA-mRNA network was constructed by using the miRDB, miRTarBase, and TargetScan databases. TFs prediction of the miRNAs in the miRNA-mRNA network was performed using the base of the TransmiR v2.0 database. Finally, a TFs-miRNA-mRNA network contributing to the onset and progression of IPF was successfully established. GO and KEGG analyses of the TFs-miRNA-mRNA network were conducted using the clusterprofiler R package. This study credibly discriminated human IPF-related miRNAs and provided a novel approach to identifying the pathological mechanisms and potential targets of IPF. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6161/rc).
Methods
Data download
First, we searched the GEO database (https://www.ncbi.nlm.nih.gov/geoprofiles) for datasets related to miRNA and IPF by using the keywords “miRNA” and “idiopathic pulmonary fibrosis”. Next, three datasets (GSE13316, GSE110147, and GSE53845) were downloaded. For miRNA expression profiling related to IPF (GSE13316, platform: GPL6955 Agilent-016436 Human miRNA Microarray 1.0), 10 samples were obtained from the surgical remnants of biopsies or lungs explanted from patients with IPF who underwent pulmonary transplant, and 10 control normal lung tissues obtained from the disease-free margins with normal histology of lung cancer resection specimens. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
For mRNA expression profiling in IPF, 22 fresh frozen lung samples were obtained from the recipient’s organs of 22 patients with IPF and 11 normal lung tissue (n=11) samples were obtained from the tissue flanking lung cancer resections (GSE110147, GPL6244 Affymetrix Human Gene 1.0 ST Array). In the GSE53845 dataset (platform: GPL6480 Agilent-014850 Whole Human Genome Microarray 4x44K G4112F), RNA was extracted directly from lung tissue samples from 40 IPF patients and eight healthy controls. The workflow of this study is shown in Figure 1.

DEMs and DEGs screening
Firstly, the DEMs between the IPF and normal tissues were screened using the GEO2R online software. P<0.05 was considered statistically significant. At the same time, the differentially expressed mRNAs between the IPF and normal tissues were also screened by the GEO2R online software. We used the Benjamini & Hochberg (false discovery rate) method to adjust the P values, and an adjusted P value <0.05 was selected as the threshold. Next, the overlapped differentially expressed mRNAs in the GSE110147 and GSE53845 datasets were selected as DEGs.
GSEA analysis of DEGs
To forecast the possible biological functions and pathways of these DEGs, GSEA was conducted using the clusterprofiler R package. P<0.05 was considered statistically significant.
WGCNA analysis and functional enrichment analysis
To screen an important module in DEGs, the GSE53845 microarray dataset was downloaded, and WGCNA analysis of DEGs was performed using the “WGCNA” R package. Pearson correlation was chosen to construct the network. We selected a soft-thresholding power of 8 when 0.8 was used as the correlation coefficient threshold. Also, the minimum number of module genes was 10, and the cutting height is 0.91. Next, the important module genes were used for GO analysis and KEGG pathway analysis. Here, an adjusted P value <0.05 was considered statistically significant.
TFs-miRNA-mRNA network construction
It is known that miRNA can bind to targeted mRNA to promote the degradation of mRNA. Herein, the targeted mRNAs of these miRNA signatures were obtained using the miRDB (http://www.mirdb.org), miRTarBase (https://mirtarbase.cuhk.edu.cn), and TargetScan (http://www.targetscan.org) databases. MRNAs presented in all three databases were regarded as targeted mRNAs of these miRNAs. By comparing the predicted targeted mRNAs with the DEGs, only the remaining overlapped mRNAs and their interaction miRNA-mRNA pairs were used to construct the miRNA-mRNA network.
Also, TFs can regulate the transcriptional expression of various RNAs. Therefore, TFs prediction of miRNAs in the miRNA-mRNA network was performed based on the TransmiR v2.0 database. Finally, an IPF-related TFs-miRNA-mRNA network was constructed. The predicted TFs were used for experimental verification.
GO and KEGG enrichment analysis of the TFs-miRNA-mRNA network
To forecast the possible functions and pathways of this TFs-miRNA-mRNA network, GO and KEGG analyses were conducted using the clusterprofiler R package. P<0.05 was considered statistically significant.
Cell culture and treatment
Human embryonic lung fibroblasts (MRC-5) was purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China). For maintenance, the cells (MRC-5) were maintained in the appropriate medium [Dulbeccos Modified Eagle Medium (DMEM)] containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco) at 37 ℃ with 5% carbon dioxide (CO2). Before determining the concentration, we searched lots of literature and found many studies (29-31) finally determined 5 ng/mL TGF-β1 as the optimal concentration by setting different concentration and comparing the effect of promoting fibrosis. In the pre-experiment we also set different concentration of TGF-β1 (2, 5, 10 ng/mL), and found when the concentration of TGF-β1 is 5 ng/mL, the effect of promoting fibrosis is the most significant. Therefore, fibroblasts (MRC-5) were cultured in a medium supplemented with 5 ng/mL recombinant TGF-β1 (Peprotech, New Jersey, USA) for 0, 24, and 48 h to induce fibroblast activation.
Verification of the candidate biomarkers
Firstly, total RNA was isolated from fibroblasts (MRC-5) using Trizol reagent (AG) and reverse-transcribed into complementary DNA (cDNA) using the Evo M-MLV RT Premix for qPCR kit (Accurate Biotechnology, Hunan, China). Then, the quantitative real time polymerase chain reaction (qRT-PCR) was carried out using the SYBR® Green Premix Pro Taq HS qPCR Kit (Rox Plus, Hunan, China) on the Agilent Mx 3000P machine (Agilent Technologies, California, USA) according to the manufacturer’s instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control for the detection. All primers in this study are shown in Table S1. The expression levels of target TFs (relative to GAPDH) were analyzed using the 2−ΔΔCt method.
Statistical analysis
All experiments were repeated at least in three times. Multiple group comparisons were assessed using a two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. All statistical analysis was done using GraphPad Prism 9 and P<0.05 was considered significant.
Results
DEMs and DEGs screening
The data in this study were analyzed separately (Figure 2). For DEMs, there were 53 differentially expressed miRNAs (30 up-regulated and 23 down-regulated) in the GSE13316 dataset. As for the DEGs, there were 15,338 differentially expressed mRNAs (6,017 up-regulated and 9,321 down-regulated) in the GSE110147 dataset, while there were 5,490 differentially expressed mRNAs (2,650 up-regulated and 2,840 down-regulated) in the GSE53845 dataset. Herein, 2,630 DEGs (1,089 up-regulated and 1,541 down-regulated) were screened. The DEMs and DEGs were used in the subsequent analyses (Figure 3A).


GSEA analysis of DEGs
As shown in Figure 3B, the GSEA analysis results indicated that these DEGs were mainly enriched in protein digestion and absorption, (extracellular matrix) ECM-receptor interaction, rheumatoid arthritis, and focal adhesion. Herein, the up-regulated genes in IPF were mainly associated with protein digestion and absorption, the phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) signaling pathway, and focal adhesion, while the down-regulated genes in IPF were mainly associated with neuroactive ligand-receptor interaction and arachidonic acid metabolism.
WGCNA and functional enrichment analysis
We selected a soft-thresholding power of 8 when 0.8 was used as the correlation coefficient threshold (Figure 4A). Through WGCNA (2,630 genes), eight co-expression modules were constructed (Figure 4B), which were independent of other modules. The blue module demonstrated the highest number of genes and significant cross-clustering in the network heatmap plot.
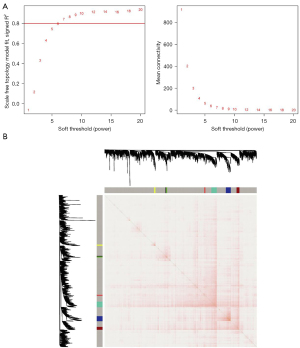
The blue module genes (86 genes) were used for the subsequent analysis (Tables S2,S3). GO functional annotation included three sub-ontologies, namely biological process (BP), cellular component (CC), and molecular function (MF). As shown in Figure 5A, DEGs in the BP category were mainly associated with cilium assembly, cilium organization, and homophilic cell adhesion via plasma membrane adhesion molecules. As for the CC terms, the DEGs were mainly involved in the ciliary basal body, plasma membrane-bounded cell projection cytoplasm, and cytoplasmic region. Regarding MF, the DEGs were mainly enriched in alpha-tubulin binding, glutathione transferase activity, and integrin binding. As shown in Figure 5B, the KEGG pathways of the DEGs were significantly enriched in fluid shear stress and atherosclerosis, drug metabolism-cytochrome P450, xenobiotic metabolism by cytochrome P450, transcriptional dysregulation in cancer, and the interleukin (IL)-17 signaling pathway.

TFs-miRNA-mRNA network
Using the miRDB, TargetScan, and miRTarBase databases, we took advantage of 53 DEMs for the second prediction and obtained 347 mRNAs. By overlapping these 347 predicted mRNAs and the previously screened 2,630 DEGs (1,089 up-regulated and 1,541 down-regulated), we obtained 11 miRNAs (hsa-miR-520b, hsa-miR-30a-5p, hsa-miR-557, hsa-miR-198, etc.) and 60 mRNAs (GALNT1, NDEL1, SETD3, GLO1, etc.).
As illustrated in Figure 6A, a TFs-miRNA-mRNA network, including 25 TFs (such as AR, ARNTL, and CEBPA), 11 miRNAs (hsa-miR-520b, hsa-miR-30a-5p, hsa-miR-557, hsa-miR-198, etc.), and 60 mRNAs (GALNT1, NDEL1, SETD3, GLO1, etc.), was constructed. This network suggested that these miRNAs or TFs might regulate the expression of some mRNAs in IPF, thereby playing critical roles in the genesis and development of IPF.
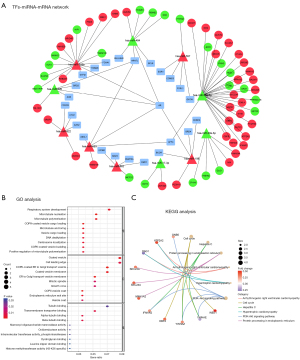
Functional enrichment analysis of the TFs-miRNA-mRNA network
To better understand this TFs-miRNA-mRNA network, GO functional annotation and KEGG pathway enrichment analyses were performed. GO functional annotation included three categories, namely BP, CC, and MF. The top 30 enriched GO items are listed in Figure 6B. The BP terms were significantly enriched in respiratory system development, microtubule nucleation, and microtubule polymerization. The CC terms were enriched in the coated vesicle, cell leading edge, and coat protein II (COPII)-coated endoplasmic reticulum (ER) to Golgi transport vesicle. The MF terms included tubulin binding and transmembrane transporter binding.
The six most important KEGG pathways are shown in Figure 6C, which included the cell cycle, the PI3K-Akt signaling pathway, hepatitis C, protein processing in the endoplasmic reticulum, arrhythmogenic right ventricular cardiomyopathy, and hypertrophic cardiomyopathy.
Experimental verification
TGF-β1 is the most important pro-fibrotic factor and plays a key role in the pathogenesis of IPF (2,10). TGF-β1-treated fibroblasts have been used by various teams to perform in vitro phenotype-related studies of lung fibrosis (31-33). Hence, in the current study, validation experiments using TGF-β1 treatment were chosen. We used qRT-PCR to verify the TFs in the TFs-miRNA-mRNA network, and the experimental results are shown in Figure 7. Following TGF-β1 treatment, ARNTL, TRIM28, EZH2, BCOR, and ASXL1 were statistically sufficiently up-regulated in a time-dependent manner. In addition, BCL6, BHLHE40, FOXA1, and EGR1 were statistically significantly down-regulated. Meanwhile, CTCF, FOSL1, AR, and FOXO3 were markedly up-regulated under TGF-β1 treatment in the 24 h groups but not in the 48 h groups.

Discussion
IPF is a chronic, progressive, fibrotic interstitial lung disease of unknown etiology that is common in elderly individuals and typically presents with characteristic imaging and histological findings (2). The psychological, physical, and socio-economic burden of IPF is substantial. With the evolution of diagnostic methods and the aging population worldwide, the prevalence and incidence rates of IPF may increase over time (34,35). The median survival of IPF patients is 2–3 years if left treated (34), and the current treatment methods are not yet satisfactory. Therefore, efforts have been made to investigate the mechanisms of IPF to discover promising therapies.
IPF is the most common type of idiopathic interstitial pneumonia, and its pathogenesis involves the dysregulation of gene networks, including both non-coding RNAs and protein-coding genes (32,36,37). Non-coding RNA lacks protein-coding ability but can still regulate a variety of cell BPs, including those related to tumorigenesis and development, aging, and the pathogenesis of IPF. In previous studies (38-40), miR-34a was found to be elevated in IPF tissues and inhibit cellular senescence through SIRT1 in Alveolar epithelial cells (AECs) and fibroblasts. Another study found that the low expression of miR-29 in IPF increases AECs antioxidants (SOD2, MnSOD, catalase) and inhibits apoptosis in AECs by regulating FOXO3A (41). Wang et al. demonstrated that long non-coding RNA SIRT-AS1 (lncRNA SIRT-AS1) can inhibit the miR-34a-mediated targeting of SIRT1 and hence suppress the progression of IPF (42). However, the comprehensive TFs-miRNA-mRNA network in IPF remains largely unknown, and the construction of this network will provide novel insights into the underlying molecular mechanisms of IPF.
In the present study, we analyzed the IPF-associated datasets from the GEO database. DEGs were identified between the IPF and normal lung tissues from the GSE110147 and GSE53845 datasets, and DEMs were identified between the IPF and normal lung tissues from the GSE13316 dataset. A Venn plot was used to obtain the intersection of DEGs. Through our analysis, 53 DEMs (30 up and 23 down) and 2,630 DEGs (1,089 up and 1,541 down) were screened. GSEA was performed to explore the possible enriched gene sets of these DEGs. These genes were related to arachidonic acid metabolism, herpes simplex virus 1 infection, the TNF signaling pathway, the IL-17 signaling pathway, neuroactive ligand-receptor interaction, the PI3K-Akt signaling pathway, protein digestion and absorption, ECM-receptor interaction, rheumatoid arthritis, and focal adhesion. It was reported previously that the anti-hepatic fibrosis effect of curcumol was related to its regulation of arachidonic acid metabolism (43). However, whether arachidonic acid metabolism is involved in the progression of IPF requires further study. As a famous immune checkpoint marker, programmed death-1 (PD-1) is upregulated in Cluster of Differentiation 4 (CD4+) T cells of IPF tissues, and increases the expression of the TF, signal transducer and activator of transcription 3 (STAT3), thereby accelerating the progression of pulmonary fibrosis by promoting the production of IL-17A and TGF-β. Our results are consistent with the previous findings, which both highlight the important role of IL-17 signaling in IPF (44).
Further, WGCNA showed that these DEGs were divided into eight modules. Turquoise module genes were utilized for GO and KEGG pathway analyses. The BP module of GO analysis showed that the blue module genes were mainly enriched in cilium-associated processes and cell adhesion, including cilium assembly, cilium organization, axoneme assembly, homophilic cell adhesion via plasma membrane adhesion molecules, cell-cell adhesion mediator activity, and myoblast migration. Interestingly, the KEGG pathway analysis showed that these genes were mainly enriched in the metabolic pathway. Glutathione metabolism, drug metabolism, and the IL-17 signaling pathway were among the top 10 pathways. Cilium-associated genes were reported to define the molecular subtypes of IPF and take part in regulating the progression of IPF (45,46). Also, glutathione S-transferases (GSTs) are reportedly enriched in pulmonary fibrosis cells and mice models. TLK117, a GSTs inhibitor, can dampen the severity of pulmonary fibrosis (47).
We used the miRDB, miRTarBase, and TargetScan databases to predict the downstream targeted mRNAs, which were then compared with DEGs to identify the overlapping mRNAs. The remaining mRNAs were applied to construct the miRNA-mRNA network. To explore the upstream TFs regulating the transcriptional expression of miRNAs, we used the TransmiR v2.0 database to build the TFs-miRNA-mRNA network. Hub miRNAs in the network included six up-regulated DEMs (hsa-miR-520b, hsa-miR-557, hsa-miR-198, hsa-miR-662, hsa-miR-623, and hsa-miR-622) and five downregulated DEMs (hsa-miR-484, hsa-miR-30a-5p, hsa-miR-30a-3p, hsa-miR-17-3p, and hsa-miR-326). Among these hub miRNAs, has-miR-30a-5p had the maximum branches and nodes, indicating that it may play an important role in the regulation of pulmonary fibrosis. Mao et al. found that miR-30a inhibits tet methylcytosine dioxygenase 1 (TET1) expression via base pairing with complementary sites in the 3'-untranslated region (3'UTR) to modulate dynamin-related protein-1 (Drp-1)-promoter hydroxymethylation. Hence, miR-30a acts as a potential therapeutic target of IPF by blocking mitochondrial fission, which is dependent on Drp-1 (48).
MicroRNA 17-92 (MiR17-92) is down-regulated in lung fibroblasts of IPF tissues compared with control tissues; the DNA methylation degree of its promotor and DNA methyltransferase 1 (DNMT1) expression increase synchronously. There is also a feedback loop between DNMT1 and microRNA 17-92 (miR17-92) in lung fibrosis (49). Das et al. demonstrated that miR-326 can down-regulate profibrotic genes such as SMAD family member 3 (Smad3), matrix metallopeptidase 9 (MMP9), and ETS proto-oncogene 1 (Ets1) and increase the expression of antifibrotic genes such as Smad7 (50). The significant decrease in miR-484 in the lung tissue or plasma of bleomycin-administered mice suggests that it could be a possible indicator of drug-induced pulmonary fibrosis (51). Our results are consistent with these previous findings, and we also found that there are limited studies on miR-520b, miR-557, miR-198, miR-662, miR-623, and miR-622 in the pathogenesis of IPF.
The network indicates that common upstream TFs, such as the androgen receptor (AR), may regulate hub miRNAs. Androgen and ARs are regulators of macrophages and monocytes in diseased lungs, such as those in asthma, chronic obstructive pulmonary disease, and lung cancer (52). To our knowledge, IPF is a lung disease that manifests sex differences (53), with higher incidence and severity in males (54-56). Additionally, the expression of ARs is greater in macrophages in males than in females (57). In the current study, AR was statistically significantly up-regulated under TGF-β1 treatment in the 24 h groups, but not in the 48 h groups. However, advanced research is required to determine whether AR is an important regulator in IPF pathogenesis. Another common TF is aryl hydrocarbon receptor nuclear translocator like (ARNTL), which is a transcriptional activator of the molecular clock feedback network that is involved in the modulation of generating circadian rhythms, cell proliferation, autophagy, and cancer cell invasion. Dong et al. showed that downregulating the expression of ARNTL significantly inhibited the canonical TGF-β1 signaling pathway and modulated TGF-β1-induced epithelial-mesenchymal transition in lung epithelial cells (58). Enhancer of zeste homolog 2 (EZH2) is the key catalytic subunit of polycomb repressive complex 2 (PRC2), which can regulate downstream gene expression via trimethylation of H3K27me3 (trimethylation at lysine 27 of histone H3). The interplay between the epigenetic regulators EZH2 and G9a is reportedly responsible for suppressing the expression of the antifibrotic factors, cytochrome c oxidase subunit II (COX2) and C-X-C motif chemokine ligand 10 (CXCL10), in IPF via H3K27me3 and H3K9me3 (59,60). Another study reported that treatment with EZH2 inhibitors could suppress M1 macrophages while promoting M2 macrophage differentiation by activating peroxisome proliferator-activated receptor (PPAR)-γ and modulating the signal transducer and activator of transcription/ suppressor of cytokine signaling (STAT/SOCS) pathway (61). MicroRNA-224 (miR-224) and its target forkhead box A1 (FOXA1) inhibit the expression of EMT markers and the migration and invasion of human lung fibroblasts in IPF under hypoxia (62). Bromodomain containing 4 (BRD4) plays a critical role in promoting pulmonary myofibroblast activation and redox imbalance, and BRD4 inhibitors is capable of inhibiting the profibrotic effects of IPF and attenuates bleomycin-induced lung fibrosis in mice (63,64). E1A binding protein p300 (EP300) is another important transcription factor in the network. In bronchial epithelial cells of IPF, EP300 were up-regulated and involved in the regulation of autophagy, cellular response to organonitrogen compounds, and collagen metabolic pathways (65). The role of forkhead box O3 (FOXO3) in pulmonary fibrosis is controversial. Qian et al. reported that overexpression of FOXO3 could reverse the antifibrosis effects induced by Angelica sinensis polysaccharide treatment (66). Lee et al. found that chitinase 1 (CHIT1) could regulate pulmonary fibrosis by modulating the TGF-β1/SMAD7 axis via interaction with FOXO3 and TGF-β receptor-associated protein 1 (TGFBRAP1) (67). However, another researcher found that FOXO3 was less expressed relative to the non-diseased controls. The knockout of FOXO3 displayed increased susceptibility to bleomycin challenge, with augmented fibrosis and higher mortality in mice (68).
MRNAs in the network were selected for GO analysis. Among the top 10 BPs, mitosis-related pathways accounted for half. A genetic study involving 1,725 IPF patients and 23,509 controls found heterozygous deleterious variants in kinesin family member 15 (KIF15), which take part in spindle separation during mitosis (69). Mitotic imbalance may be an important mechanism in the pathogenesis of IPF. The six most important pathways analyzed by KEGG included the cell cycle, the PI3K-Akt signaling pathway, hepatitis C, protein processing in the endoplasmic reticulum, arrhythmogenic right ventricular cardiomyopathy, and hypertrophic cardiomyopathy. Numerous reports have indicated that the PI3K-Akt signaling pathway plays an important role in the pulmonary fibrosis process. In recent years, various research teams have utilized PI3K/AKT pathway inhibitors in animal models (70) and clinical trials (71) to investigate their effects on the process of pulmonary fibrosis, and they have achieved important results. The administration of a pathway inhibitor in a mouse model of IPF was shown to significantly inhibit PI3K activation and suppress the production of hydroxyproline, thereby reducing collagen deposition and ultimately improving mouse survival.
Our research still has certain limitations that should be noted. Firstly, the data used in this study came from the GEO public database; for retrospective analysis, this level of evidence is insufficient. Secondly, the regulatory network lacks expression verification in in vivo models, and thus, in-depth mechanism research should be conducted after verifying the definite expression correlation.
Conclusions
The novel TFs-miRNA-mRNA network that we constructed could provide new insights into the underlying molecular mechanisms of IPF. ARNTL, TRIM28, EZH2, BCOR, ASXL1, BCL6, BHLHE40, FOXA1, and EGR1 may play important roles in IPF and become effective biomarkers for diagnosis and treatment.
Acknowledgments
The results shown in the current study are mainly based on data obtained from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/).
Funding: This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81902559), the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2019A1515110787), and the Dean’s Foundation of Zhujiang Hospital of Southern Medical University (Grant Nos. yzjj2018rc08 and yzjj2021qn14).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6161/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6161/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6161/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788-824. [Crossref] [PubMed]
- Lederer DJ, Martinez FJ. Idiopathic Pulmonary Fibrosis. N Engl J Med 2018;378:1811-23. [Crossref] [PubMed]
- Vancheri C, Failla M, Crimi N, et al. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J 2010;35:496-504. [Crossref] [PubMed]
- Natsuizaka M, Chiba H, Kuronuma K, et al. Epidemiologic survey of Japanese patients with idiopathic pulmonary fibrosis and investigation of ethnic differences. Am J Respir Crit Care Med 2014;190:773-9. [Crossref] [PubMed]
- Navaratnam V, Fleming KM, West J, et al. The rising incidence of idiopathic pulmonary fibrosis in the U.K. Thorax 2011;66:462-7. [Crossref] [PubMed]
- Nalysnyk L, Cid-Ruzafa J, Rotella P, et al. Incidence and prevalence of idiopathic pulmonary fibrosis: review of the literature. Eur Respir Rev 2012;21:355-61. [Crossref] [PubMed]
- Hutchinson J, Fogarty A, Hubbard R, et al. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur Respir J 2015;46:795-806. [Crossref] [PubMed]
- Selman M, King TE, Pardo A, et al. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:136-51. [Crossref] [PubMed]
- Raghu G, Anstrom KJ, et al. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med 2012;366:1968-77. [Crossref] [PubMed]
- Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017;389:1941-52. [Crossref] [PubMed]
- Tian L, Cao J, Deng X, et al. Unveiling transcription factor regulation and differential co-expression genes in Duchenne muscular dystrophy. Diagn Pathol 2014;9:210. [Crossref] [PubMed]
- Bhattacharyya S, Wu M, Fang F, et al. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol 2011;30:235-42. [Crossref] [PubMed]
- Cadena-Suárez AR, Hernández-Hernández HA, Alvarado-Vásquez N, et al. Role of MicroRNAs in Signaling Pathways Associated with the Pathogenesis of Idiopathic Pulmonary Fibrosis: A Focus on Epithelial-Mesenchymal Transition. Int J Mol Sci 2022;23:6613. [Crossref] [PubMed]
- Ye Z, Hu Y. TGF-β1: Gentlemanly orchestrator in idiopathic pulmonary fibrosis Int J Mol Med 2021;48:132. (Review). [Crossref] [PubMed]
- Huang Y, Xie Y, Abel PW, et al. TGF-β1-induced miR-424 promotes pulmonary myofibroblast differentiation by targeting Slit2 protein expression. Biochem Pharmacol 2020;180:114172. [Crossref] [PubMed]
- Deng X, Jin K, Li Y, et al. Platelet-Derived Growth Factor and Transforming Growth Factor β1 Regulate ARDS-Associated Lung Fibrosis Through Distinct Signaling Pathways. Cell Physiol Biochem 2015;36:937-46. [Crossref] [PubMed]
- Kadoya K, Togo S, Tulafu M, et al. Specific Features of Fibrotic Lung Fibroblasts Highly Sensitive to Fibrotic Processes Mediated via TGF-β-ERK5 Interaction. Cell Physiol Biochem 2019;52:822-37. [Crossref] [PubMed]
- Ghatak S, Markwald RR, Hascall VC, et al. Transforming growth factor β1 (TGFβ1) regulates CD44V6 expression and activity through extracellular signal-regulated kinase (ERK)-induced EGR1 in pulmonary fibrogenic fibroblasts. J Biol Chem 2017;292:10465-89. [Crossref] [PubMed]
- Pandit KV, Corcoran D, Yousef H, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;182:220-9. [Crossref] [PubMed]
- Strell C, Norberg KJ, Mezheyeuski A, et al. Stroma-regulated HMGA2 is an independent prognostic marker in PDAC and AAC. Br J Cancer 2017;117:65-77. [Crossref] [PubMed]
- Thuault S, Valcourt U, Petersen M, et al. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol 2006;174:175-83. [Crossref] [PubMed]
- Liu G, Friggeri A, Yang Y, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010;207:1589-97. [Crossref] [PubMed]
- Yang S, Banerjee S, de Freitas A, et al. Participation of miR-200 in pulmonary fibrosis. Am J Pathol 2012;180:484-93. [Crossref] [PubMed]
- Zhu K, Xu A, Xia W, et al. Integrated analysis of the molecular mechanisms in idiopathic pulmonary fibrosis. Int J Med Sci 2021;18:3412-24. [Crossref] [PubMed]
- Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Sato Y, et al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49:D545-51. [Crossref] [PubMed]
- Gu L, Zhu YJ, Yang X, et al. Effect of TGF-beta/Smad signaling pathway on lung myofibroblast differentiation. Acta Pharmacol Sin 2007;28:382-91. [Crossref] [PubMed]
- Zhang Q, Tu W, Tian K, et al. Sirtuin 6 inhibits myofibroblast differentiation via inactivating transforming growth factor-β1/Smad2 and nuclear factor-κB signaling pathways in human fetal lung fibroblasts. J Cell Biochem 2019;120:93-104. [Crossref] [PubMed]
- Xu Q, Cheng D, Li G, et al. CircHIPK3 regulates pulmonary fibrosis by facilitating glycolysis in miR-30a-3p/FOXK2-dependent manner. Int J Biol Sci 2021;17:2294-307. [Crossref] [PubMed]
- Li J, Li P, Zhang G, et al. CircRNA TADA2A relieves idiopathic pulmonary fibrosis by inhibiting proliferation and activation of fibroblasts. Cell Death Dis 2020;11:553. [Crossref] [PubMed]
- Liu P, Luo G, Dodson M, et al. The NRF2-LOC344887 signaling axis suppresses pulmonary fibrosis. Redox Biol 2021;38:101766. [Crossref] [PubMed]
- Maher TM, Bendstrup E, Dron L, et al. Global incidence and prevalence of idiopathic pulmonary fibrosis. Respir Res 2021;22:197. [Crossref] [PubMed]
- Diamantopoulos A, Wright E, Vlahopoulou K, et al. The Burden of Illness of Idiopathic Pulmonary Fibrosis: A Comprehensive Evidence Review. Pharmacoeconomics 2018;36:779-807. [Crossref] [PubMed]
- Zhou J, Lin Y, Kang X, et al. microRNA-186 in extracellular vesicles from bone marrow mesenchymal stem cells alleviates idiopathic pulmonary fibrosis via interaction with SOX4 and DKK1. Stem Cell Res Ther 2021;12:96. [Crossref] [PubMed]
- Yi H, Luo D, Xiao Y, Jiang D. Knockdown of long non-coding RNA DLEU2 suppresses idiopathic pulmonary fibrosis by regulating the microRNA-369-3p/TRIM2 axis. Int J Mol Med 2021;47:80. [Crossref] [PubMed]
- Cui H, Ge J, Xie N, et al. miR-34a promotes fibrosis in aged lungs by inducing alveolarepithelial dysfunctions. Am J Physiol Lung Cell Mol Physiol 2017;312:L415-24. [Crossref] [PubMed]
- Cui H, Ge J, Xie N, et al. miR-34a Inhibits Lung Fibrosis by Inducing Lung Fibroblast Senescence. Am J Respir Cell Mol Biol 2017;56:168-78. [Crossref] [PubMed]
- Disayabutr S, Kim EK, Cha SI, et al. miR-34 miRNAs Regulate Cellular Senescence in Type II Alveolar Epithelial Cells of Patients with Idiopathic Pulmonary Fibrosis. PLoS One 2016;11:e0158367. [Crossref] [PubMed]
- Xie T, Liang J, Geng Y, et al. MicroRNA-29c Prevents Pulmonary Fibrosis by Regulating Epithelial Cell Renewal and Apoptosis. Am J Respir Cell Mol Biol 2017;57:721-32. [Crossref] [PubMed]
- Wang Y, Pang WJ, Wei N, et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene 2014;539:117-24. [Crossref] [PubMed]
- Zheng Y, Wang J, Wang J, et al. Gut microbiota combined with metabolomics reveal the mechanism of curcumol on liver fibrosis in mice. Biomed Pharmacother 2022;152:113204. [Crossref] [PubMed]
- Celada LJ, Kropski JA, Herazo-Maya JD, et al. PD-1 up-regulation on CD4(+) T cells promotes pulmonary fibrosis through STAT3-mediated IL-17A and TGF-β1 production. Sci Transl Med 2018;10:eaar8356. [Crossref] [PubMed]
- McDonough JE, Kaminski N, Thienpont B, et al. Gene correlation network analysis to identify regulatory factors in idiopathic pulmonary fibrosis. Thorax 2019;74:132-40. [Crossref] [PubMed]
- Yang IV, Coldren CD, Leach SM, et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax 2013;68:1114-21. [Crossref] [PubMed]
- He N, Bai S, Huang Y, et al. Evaluation of Glutathione S-Transferase Inhibition Effects on Idiopathic Pulmonary Fibrosis Therapy with a Near-Infrared Fluorescent Probe in Cell and Mice Models. Anal Chem 2019;91:5424-32. [Crossref] [PubMed]
- Mao C, Zhang J, Lin S, et al. MiRNA-30a inhibits AECs-II apoptosis by blocking mitochondrial fission dependent on Drp-1. J Cell Mol Med 2014;18:2404-16. [Crossref] [PubMed]
- Dakhlallah D, Batte K, Wang Y, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013;187:397-405. [Crossref] [PubMed]
- Das S, Kumar M, Negi V, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol 2014;50:882-92. [Crossref] [PubMed]
- Konaka T, Kawami M, Yamamoto A, et al. miR-484: A Possible Indicator of Drug-Induced Pulmonary Fibrosis. J Pharm Pharm Sci 2020;23:486-95. [Crossref] [PubMed]
- Becerra-Diaz M, Song M, Heller N. Androgen and Androgen Receptors as Regulators of Monocyte and Macrophage Biology in the Healthy and Diseased Lung. Front Immunol 2020;11:1698. [Crossref] [PubMed]
- Han MK, Murray S, Fell CD, et al. Sex differences in physiological progression of idiopathic pulmonary fibrosis. Eur Respir J 2008;31:1183-8. [Crossref] [PubMed]
- Mortimer KM, Bartels DB, Hartmann N, et al. Characterizing Health Outcomes in Idiopathic Pulmonary Fibrosis using US Health Claims Data. Respiration 2020;99:108-18. [Crossref] [PubMed]
- Olson AL, Swigris JJ, Lezotte DC, et al. Mortality from pulmonary fibrosis increased in the United States from 1992 to 2003. Am J Respir Crit Care Med 2007;176:277-84. [Crossref] [PubMed]
- Gribbin J, Hubbard RB, Le Jeune I, et al. Incidence and mortality of idiopathic pulmonary fibrosis and sarcoidosis in the UK. Thorax 2006;61:980-5. [Crossref] [PubMed]
- McCrohon JA, Death AK, Nakhla S, et al. Androgen receptor expression is greater in macrophages from male than from female donors. A sex difference with implications for atherogenesis. Circulation 2000;101:224-6. [Crossref] [PubMed]
- Dong C, Gongora R, Sosulski ML, et al. Regulation of transforming growth factor-beta1 (TGF-β1)-induced pro-fibrotic activities by circadian clock gene BMAL1. Respir Res 2016;17:4. [Crossref] [PubMed]
- Coward WR, Brand OJ, Pasini A, et al. Interplay between EZH2 and G9a Regulates CXCL10 Gene Repression in Idiopathic Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2018;58:449-60. [Crossref] [PubMed]
- Coward WR, Feghali-Bostwick CA, Jenkins G, et al. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J 2014;28:3183-96. [Crossref] [PubMed]
- Bao X, Liu X, Liu N, et al. Inhibition of EZH2 prevents acute respiratory distress syndrome (ARDS)-associated pulmonary fibrosis by regulating the macrophage polarization phenotype. Respir Res 2021;22:194. [Crossref] [PubMed]
- Jeong SH, Son ES, Lee YE, et al. Histone deacetylase 3 promotes alveolar epithelial-mesenchymal transition and fibroblast migration under hypoxic conditions. Exp Mol Med 2022;54:922-31. [Crossref] [PubMed]
- Tang X, Peng R, Phillips JE, et al. Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am J Pathol 2013;183:470-9. [Crossref] [PubMed]
- Stock CJW, Michaeloudes C, Leoni P, et al. Bromodomain and Extraterminal (BET) Protein Inhibition Restores Redox Balance and Inhibits Myofibroblast Activation. Biomed Res Int 2019;2019:1484736. [Crossref] [PubMed]
- Tao J, Zhang M, Wen Z, et al. Inhibition of EP300 and DDR1 synergistically alleviates pulmonary fibrosis in vitro and in vivo. Biomed Pharmacother 2018;106:1727-33. [Crossref] [PubMed]
- Qian W, Cai X, Qian Q, et al. Angelica Sinensis Polysaccharide Suppresses Epithelial-Mesenchymal Transition and Pulmonary Fibrosis via a DANCR/AUF-1/FOXO3 Regulatory Axis. Aging Dis 2020;11:17-30. [Crossref] [PubMed]
- Lee CM, He CH, Park JW, et al. Chitinase 1 regulates pulmonary fibrosis by modulating TGF-β/SMAD7 pathway via TGFBRAP1 and FOXO3. Life Sci Alliance 2019;2:e201900350. [Crossref] [PubMed]
- Al-Tamari HM, Dabral S, Schmall A, et al. FoxO3 an important player in fibrogenesis and therapeutic target for idiopathic pulmonary fibrosis. EMBO Mol Med 2018;10:276-93. [Crossref] [PubMed]
- Zhang D, Povysil G, Kobeissy PH, et al. Rare and Common Variants in KIF15 Contribute to Genetic Risk of Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 2022;206:56-69. [Crossref] [PubMed]
- Hettiarachchi SU, Li YH, Roy J, et al. Targeted inhibition of PI3 kinase/mTOR specifically in fibrotic lung fibroblasts suppresses pulmonary fibrosis in experimental models. Sci Transl Med 2020;12:eaay3724. [Crossref] [PubMed]
- Lukey PT, Harrison SA, Yang S, et al. A randomised, placebo-controlled study of omipalisib (PI3K/mTOR) in idiopathic pulmonary fibrosis. Eur Respir J 2019;53:1801992. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


