
MiR-21-5p protects against ischemic stroke by targeting IL-6R
Highlight box
Key findings
• Hsa-miR-21-5p can inhibit IL-6R expression.
What is known and what is new?
• MiRNAs have the capacity to relieve or rescue the ischemia damage.
• Hsa-miR-21-5p was found to be critical in ischemic stroke by differential analysis of the GSE158313 dataset and clinical trial analysis.
What is the implication, and what should change now?
• Hsa-miR-21-5p is known to be associated with IL-6R during ischemic stroke disease through comprehensive analysis of multiple database screens.
Introduction
Ischemic stroke is a neurologic systematic disease, the major pathology of which is brain injury caused by short- or long-term vascular occlusion. There are an estimated 40,000,000 ischemic stroke patients worldwide per year, and the incidence is still elevated in recent years. In developing countries, the morbidity of ischemic stroke is increasing with a 10% growth rate each year (1). Stroke can be dived into two subtypes: “ischemic stroke” caused by cerebral blood circulation insufficiency and “hemorrhagic stroke” caused by cerebral bleeding, both of which lead to brain damage and dysfunction (2,3). Currently, therapies for ischemic stroke patients include exercise, diet control, and drug treatment, but there are no effective methods to cure patients with brain dysfunction. Due to the lack treatment options to reverse brain damage, it is extremely important to explore ischemia-related biomarkers in the early stage. The early diagnosis and treatment could effectively avoid severe complications and death (4).
MicroRNA (miRNA) is a type of short RNA containing 20–24 nucleotides, which can bind to the 3'-untranslated region (3'-UTR) of messenger RNAs (mRNAs) and block mRNAs’ transcription, and then silence gene expression. There are numerous publications showing the important roles of miRNAs in regulating biological pathways related to cellular apoptosis, autophagy, and inflammation (5,6). For example, miR-543 could bind multiple mRNAs including WNT, PTEN, BRIP1, and SIRT1, thus regulating the WNT, PI3K/Akt, and MAPK/ERK signal pathways in various cancer cells (7). In ischemic stroke brain tissues, about 20% of miRNA expression levels are significantly changed with the advancement of disease (8). Therefore, miRNAs may be the key regulator of ischemic stroke and could be a kind of potential and powerful therapeutic target.
Recent articles show that miR-21-5p harbors the protective function by reducing cellular apoptosis levels in rat coronary heart disease models and regulating PTEN/PDCD4 expression to inhibit apoptosis in mice pulmonary ischemia-reperfusion models (9,10). As an important miRNA in cell proliferation and apoptosis, numerous publications have reported that the expression levels of hsa-miR-21-5p are changed in many ischemic diseases (11-14). The miRNA hsa-miR-21-5p is located on the human chromosome 17q23.2 locus, which is nearly downstream of vacuole membrane protein 1 (VMP1) (15). In many diseases, especially cancer, hsa-miR-21-5p usually regulates the PI3K/Akt pathway by suppressing PTEN and influencing cell proliferation and death (16). However, the role of hsa-miR-21-5p in stroke is still unclear. Hence, in this study, we established an in vivo ischemia cellular model and explored the function of hsa-miR-21-5p in microvascular epithelial cells, which provides a new insight and potential target for ischemic stroke treatment in the future. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6451/rc).
Methods
GEO datasets analysis
The datasets GSE158313, GSE22255, and GSE16561 are downloaded from the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) website (17-20). Sequencing data were analyzed for the exploration of ischemia stroke-related miRNAs using the “DEGs” package in R (R Foundation for Statistical Computing, Vienna, Austria), and the significantly changed miRNAs [|log2fold change (FC)| >2, and P<0.05] were included in the following analysis. The top 30 of the expression level-changed miRNAs were plotted in the heatmap through the “HeatMap2” package. A Venn plot was drawn using the “Venn” package. The co-expression correlation coefficient of genes in R was calculated using “cor” function to verify the co-expression interaction relationship of genes. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) function enrichment analysis of target genes and “DEGs” was carried out through the “cluster profile” package of R software, and the results were analyzed and visualized through the cluster diagram generated by “org.Hs.eg.db” and “GOplot” R package.
Patient’s samples
A total of 60 ischemic stroke patients from the Second Affiliated Hospital of Qiqihar Medical University from September 2020 to January 2022 were included in this study. Peripheral blood samples were collected on the second day after patients’ admission. A total of 23 age-matched healthy people were also recruited and had their peripheral blood samples collected as the control group. All clinical participants had been informed of the content of the experiment and signed the informed consent form. The exclusion criteria were as follows: (I) the presence of ischemia in any other organs; (II) any psychiatric diseases; (III) pregnancy; (IV) liver and kidney dysfunction or autoimmune disease patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The Second Affiliated Hospital of Qiqihar Medical University (No. 2021-0507).
Cell culture
The human microvascular epithelial cell line HMEC-1 was purchased from American Type Culture Collection (ATCC), and the 293T cells were a gift from another lab. The HMEC-1 cells were cultured in MCBD 131 medium (10372-019, Gibco, Grand Island, NY, USA) containing10% fetal bovine serum (FBS; 10099141C, Gibco), 100 U/mL penicillin/streptomycin (Pen/Strep; 15140122, Gibco), 2 mM L-glutamine (A2916801, Gibco), and 0.05 mg/mL hydrocortisone (H6909, Sigma Aldrich, St. Louis, MO, USA). The 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM; 11965092, Gibco) containing 10% FBS and 100 U/mL Pen/Strep. All cells were cultivated at 37 ℃ with 5% CO2 environment.
The miRNA mimic/inhibitor and plasmids were transfected using Lipofectamine 3000 reagents (L3000015, Invitrogen, Carlsbad, CA, USA). All cells were transfected at the 70% cell fusion rate and following the manufacturer’s protocol, the cells are collected for analysis after 48 hours of transfection. The inhibitory effect of inhibitors on hsa-miR-21-5p was stronger than that of inhibitors, therefore, inhibitors will be used to inhibit hsa-miR-21-5p in the study. The miRNA mimic/inhibitor were purchased from GenePharma (Shanghai, China), the sequences are shown in Table 1.
Table 1
| ID | Sequence |
|---|---|
| Control mimic | 5'-UUCUCCGAACGUGUCACGUTT-3' |
| Hsa-miR-21-5p mimic | 5'-CAACACCAGUCGAUGGGCUGU-3' |
| Hsa-miR-21-5p inhibitor | 5'-ACAGCCCAUCGACUGGUGUUG-3' |
MiRNA, microRNA.
Oxygen-glucose deprivation (OGD) treatment
The OGD was performed to establish the in vivo ischemia-reperfusion model. For OGD-treated cells, the culture medium was changed to DMEM with non-glucose and non-FBS. Then, the cells were transferred to an environment containing 5% CO2 and 95% N2 at 37 ℃ and treated for 2-, 4-, 6-, and 8-hour following the experimental design. Next, the cells were transferred to the original culture medium and cultivated in a normal oxygen concentration environment at 37 ℃ (15).
Dual-luciferase assay
The wild-type (WT) and mutant interleukin-6 receptor (IL-6R) 3'-UTR were replicated and cloned into luciferase reporter vector pGL3 (Promega, Madison, WI, USA) respectively. Then, the hsa-miR-21-5p mimic and control mimic were co-transfected with pGL3-IL-6R 3'-UTR plasmids or pGL3-IL-6R 3'-UTR mutant into 293T cells. After 48 hours of transfection, the luciferase activities were detected through dual-luciferase reporter assay system (Promega) following the manufacturer’s protocol. Fluorescence was measured using the Promega GloMaxTM 20/20 Luminometer, and luciferase activity was normalized with the ratio of fluorescence values of firefly luciferase and Renilla luciferase (16,17).
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNAs were extracted from blood or cells using Trizol (Invitrogen) reagent. For mRNA detection, the total RNAs were reverse transcribed to complementary DNA (cDNA) by Takara reverse transcription kit (RR047A, Takara, Shiga Japan). For miRNA detection, the reverse transcription was performed using a miRNA First Strand cDNA Synthesis (Tailing Reaction) kit (B532451-0020, Sangon, Shanghai, China). QRT-PCR was performed with SYBR® Premix Ex TaqTM II (Perfect Real Time) kit (DRR081, Takara, Japan) and detected by the real-time RT-PCR system (ABI 7500, ABI, Sterling, VA, USA). The RT-PCR reaction protocol was as follows: pre-denature, 95 ℃ for 30 seconds, PCR reaction: 95 ℃ for 5 seconds, and 60 ℃ for 35 seconds, recycle 40 times. Each sample was tested with 3 repeats. The universal reverse primer for miRNA was supplied by the miRNA First Strand cDNA Synthesis (Tailing Reaction) kit (Sangon, Shanghai, China), and other primers were synthesized by Sangon, the sequence details of which are listed in Table 2. The cycle threshold (Ct) values of each sample were recorded and calculated compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 Ct values with 2−ΔΔCt methods (18,19).
Table 2
| Gene | Primer sequence |
|---|---|
| Hsa-miR-21-5p | F: 5'-ACACTCCAGCTGGGTAGCTTATCAGACTGA-3' |
| R: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACATC-3' | |
| Hsa-miR-548ar-3p | F: 5'-ACACTCCAGCTGGGTAAAACTGCAGTTAT-3' |
| R: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCAAAAAT-3' | |
| Hsa-miR-651-5p | F: 5'-ACACTCCAGCTGGGAAAGGAAAGTGTATCC-3' |
| R: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAG CTTTTAGG-3' | |
| Hsa-miR-142-5p | F: 5'-ACACTCCAGCTGGGTGTAGTGTTTCCTACTTT-3' |
| R: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAG TCCATAAA-3' | |
| Hsa-miR-30e-5p | F: 5'-ACACTCCAGCTGGGTGTAAACATCCTTGAC-3' |
| R: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAG CTTCCAGT-3' | |
| U6 | F: 5'-CTCGCTTCGGCAGCACA-3' |
| R: 5'-AACGCTTCACGAATTTGCGT-3' | |
| IL-6R | F: 5'-CCCCTCAGCAATGTTGTTTGT-3' |
| R: 5'-CTCCGGGACTGCTAACTGG-3' | |
| FBXO11 | F: 5'-GGTCATCGTGCAAAACGTGC-3' |
| R: 5'-ACAAGCTGCTCTACAAAGATCC-3' | |
| FAM46A | F: 5'-AGGGTGAAGGGTACTTCGC-3' |
| R: 5'-CTTTCGCAATAGTCCAAGCAATG-3' | |
| GAPDH | F: 5'-CGACACTTTATCATGGCTA-3' |
| R: 5'-TTGTTGCCGATCACTGAAT-3' |
RT-PCR, reverse transcription polymerase chain reaction; IL-6R, interleukin-6 receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Western blot
The cultured or treated cells were washed with warm phosphate-buffered saline (PBS) 2 times, then collected with radioimmunoprecipitation assay (RIPA) solution and sonicated for 15 seconds to disrupt the cells, and the tube was then placed on ice for 30 minutes. The mixture was centrifuged at 12,000 rpm for 20 minutes at 4 ℃ and the supernatant was collected. The protein concentrations are tested by the bicinchoninic acid (BCA) Protein Assay Kit (PC0020, Solarbio, Beijing, China). About 100 µL protein solution was taken and 5× sodium dodecyl sulfate (SDS) loading buffer was added (P1040, Solarbio, China), then the samples were boiled at 95 ℃ for 10 minutes to denature the proteins. The samples were loaded into 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels (P0052A, Beyotime, China), then transferred to a polyvinylidene fluoride (PVDF) membrane (IPVH00010, Merck, Kenilworth, NJ, USA) and blocked with 5% non-fat milk (P0216, Beyotime, China) for 1 hour at room temperature. Next, the PVDF membranes were fertilized with the diluted primary antibody including GAPDH (ab8245, 1:10,000, Abcam, Cambridge, UK), IL-6R (ab222101, 1:1,000, Abcam), Bcl-2 (ab32124, 1:1,000, Abcam), Bax (ab182734, 1:1,000, Abcam), caspase-3 (ab32351, 1:1,000, Abcam), cleaved-caspase-3 (ab32042, 1:1,000, Abcam), caspase-9 (ab32539, 1:1,000, Abcam), and cleaved-caspase-9 (ab2324, 1:1,000, Abcam) for overnight at 4 ℃. On the second day, the membranes were washed with 1× tris-buffered saline with Tween 20 (TBST) 3 times and fertilized with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 hour at room temperature. Then, the membranes were exposed with chemiluminescent HRP Substrate enhanced chemiluminescence (ECL) (WBKLS0500, Millipore, Burlington, MA, USA) and captured by the Bio-Rad ChemiDoc MP Imaging system (Bio-Rad, Hercules, CA, USA). The images were quantified using Quantity One v4.6.2 software and normalized with the grey values of GAPDH, and each experiment was repeated at least 3 times (20).
3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay
The cells are planted in a 96-well plate and cultured for 48 hours. After the treatment, the cells were added to 10 µL MTT solution (C0009S, Beyotime, China) and incubated at 37 ℃ for 4 hours, then added 100 µL formazan solution for 2 hours. The cell viabilities were detected by the optical density (OD) values at 570 nm and normalized to the control group.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
The cell apoptosis levels are detected using the One Step TUNEL Apoptosis Assay Kit (C1086, Beyotime, China) following the manufacturer’s protocol. In brief, the cultured cells were removed from the culture medium and washed with 1× PBS once, fixed with 4% paraformaldehyde (PFA) for 10 minutes then washed with 1× PBS once. The fixed cells were penetrated with 0.3% Triton X-100 in PBS for 5 minutes at room temperature. Next, washed cells with 1× PBS three times, and added 50 µL TUNEL solution fertilized at 37 ℃ for 1 hour. The TUNEL-positive cells of samples were then quantified using a microscope.
Statistical analysis
The statistical analysis was performed with SPSS 21.0 software (IBM Corp., Armonk, NY, USA), and the error bars were mean ± standard deviation (SD). Statistical significance between experimental groups was assessed using one-way analysis of variance (ANOVA), unpaired Student’s t-test, or non-parametric test. Statistical significance was defined as the P value <0.05.
Results
Hsa-miR-21-5p is decreased in the peripheral blood of ischemia stroke patients
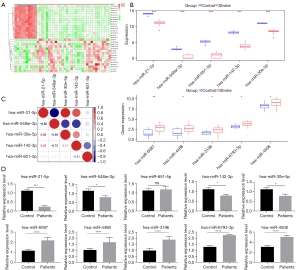
We first searched the GEO database to explore ischemia stroke-related non-coding RNAs sequencing data and find the GSE158313 dataset contained the miRNAs sequencing data of ischemia stroke patients. The GSE158313 datasets include 33 stroke patients and 10 healthy controls, the sequencing samples are the peripheral blood of healthy people and patients 2 days after stroke. Through analysis of differentially expressed miRNAs in the GSE1583131 using the “DEGs” package, we identified a cluster of miRNAs which show variant expression in patients compared to healthy people, and the TOP30 differentially expressed miRNAs were plotted in a heatmap (Figure 1A). Our results showed that hsa-miR-548ar-3p, hsa-miR-651-5p, hsa-miR-142-3p, hsa-miR-21-5p, and hsa-miR-30e-5p were most decreased miRNAs in the patients’ blood. The expression levels of has-miR-6087, has-miR-4488, has-miR-3196, has-miR-6793-3p and has-miR-4508 were up-regulated (Figure 1B). In the correlation heat map drawn by the interaction analysis of the 5 miRNAs obtained, red represents positive correlation and blue represents negative correlation (Figure 1C). We performed qRT-PCR assay to verify these 10 miRNAs expression levels in 60 ischemia stroke patients and 23 age-matched healthy control people in hospital. The qRT-PCR results showed that hsa-miR-21-5p is the most reduced miRNA in patients’ blood compared with controls (P<0.001) (Figure 1D). Thus, we chose the hsa-miR-21-5p as our focus miRNA in the subsequent experiments.
Functional enrichment analysis of target genes
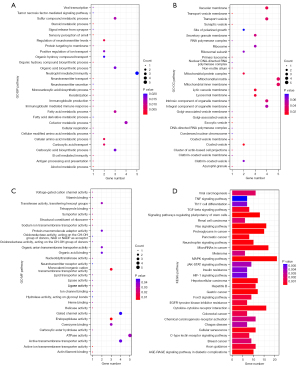
To explore the function of hsa-miR-231-5p in the ischemic stroke process, we predicted the potential target mRNA of hsa-miR-21-5p through the TargetScan website (https://www.targetscan.org/vert_80/) and retrieved a total of 384 targeted mRNAs. Functional enrichment analysis of these 384 target genes (Figure 2A-2D) illustrated the top 30 most significant GO terms and KEGG pathways.
Screening for target genes of hsa-miR-21-5p
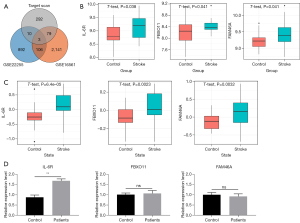
We screened the RNA sequencing (RNA-seq) data of ischemic stroke patients in the NCBI database and found that the GSE22255 and GSE16561 datasets that meet our criteria. In the GSE22255 dataset, there are 20 patients and 20 age-matched healthy people, and there are also 39 patients and 24 healthy people in the GSE16561. We uncovered 1,011 and 2,329 up-regulated genes in the GSE22255 and GSE16561 datasets, respectively, through analysis by the limma package (Figure 3A). Next, we intersected the 3 gene sets, the significantly changed genes in GSE22255, the significantly changed genes in GSE16561, and the hsa-miR-21-5p potential target genes, and ascertained 3 genes: IL-6R, FBXO11, and FAM46A (Figure 3A). In the GSE22255 and GSE16561 datasets, the IL-6R showed more elevated expression compared to FBOX11 and FAM46A in patients (Figure 3B,3C). Also, IL-6R presents notable elevated expression in the peripheral blood of stroke patients (P<0.01). FBXO11 and FAM46A showed no significant changes in patients and healthy people’s blood (both P>0.05) (Figure 3D).
Functional enrichment analysis of differentially expressed genes (DEGs)
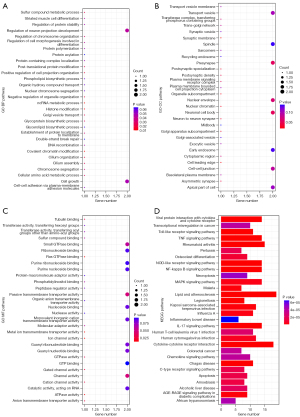
According to the median expression of IL-6R, stroke patients in the GSE22255 dataset were divided into a high expression group and a low expression group, and Bayesian t-test was performed between the two groups. According to |log2FC| >1 and P<0.05 as the cut-off value, 177 DEGs were obtained, and functional enrichment analysis was conducted according to these 177 DEGs. The results showed that these DEGs were enriched in the lipid atherosclerosis and cytokine-cytokine receptor interaction pathways, and played a role in cell apoptosis. Figure 4A-4D illustrates the top 30 most significant GO terms and KEGG pathways.
The function of hsa-miR-21-5p in ischemia-reperfusion in-vivo model
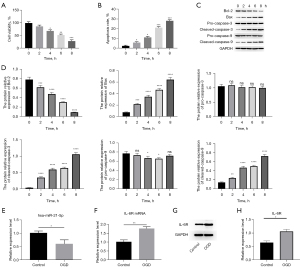
To explore the role of hsa-miR-21-5p in ischemia-reperfusion, we established the in vivo ischemia-reperfusion cell model on human microvascular epithelial cell lines HMEC-1 through OGD treatment. First, we detected the cell viabilities of OGD treated 0-, 2-, 4-, 6-, and 8-hour cells using the MTT assay kit, and the cell apoptosis levels were quantified using the TUNEL staining kit. The results showed that the cell viabilities were decreased after 4-, 6-, and 8-hour of OGD treatment, and the cell viability levels were negatively affected following treatment time (Figure 5A). The apoptosis cells were significantly increased after 2-, 4-, 6-, and 8-hour of OGD treatment, and the TUNEL-positive cells were increased with treatment time (Figure 5B). The western blotting results also showed that apoptosis-related proteins including Bax, cleaved-capsase-3, and cleaved-caspase-9 were elevated following 4-, 6-, and 8-hour of OGD treatment (P<0.05, P<0.01, P<0.001, respectively), and Bcl-2 protein are descended after 4-, 6-, and 8-hour of OGD treatment (P<0.05, P<0.01, P<0.001, respectively) (Figure 5C,5D). The above results hint that the OGD-treated HMEC-1 cells could successfully mimic the injury of ischemia-reperfusion caused by the prolonged deficit of oxygen and blood. Since the 6-hour OGD-treated cells showed significant changes of cell viability (P<0.01) (Figure 5A) and apoptosis level (P<0.01) (Figure 5B) compared to non-OGD-treated cells, the Bax (P<0.01), cleaved-caspase-3 (P<0.01), cleaved-caspase-6 (P<0.01), and Bcl-2 (P<0.01) levels were also significantly changed in the 6-hour OGD-treated group (Figure 5C,5D). Thus, we chose the 6-hour OGD-treated cells as the treatment group in the following studies. After OGD treatment, the expression level of hsa-miR-21-5p was significantly decreased in the OGD group (P<0.05) (Figure 5E). Notably, both the IL-6R mRNA and protein levels were upregulated in the OGD treatment group compared with the controls (Figure 5F-5H). In general, the hsa-miR-21-5p was reduced and IL-6R was increased in HMEC-1 cells after OGD treatment.
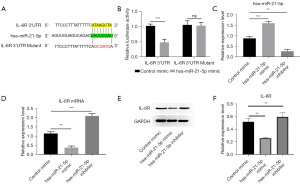
Hsa-miR-21-5p could bind IL-6R mRNA and inhibit its translation
According to the prediction of the TargetScan database, IL-6R is a potential target of hsa-miR-21-5p. To verify this prediction, we cloned the 2964-2961 site of the IL-6R 3'-UTR which is predicted to be bound into the luciferase reporter vector via hsa-miR-21-5p and constructed the mutant 3'-UTR of IL-6R into the vector. Thus, we established the IL-6R-3'-UTR and IL-6R-3'-UTR-mutant plasmids and transfected these plasmids into 293T cells (Figure 6A). The results showed that the luciferase activity of hsa-miR-21-5p + IL-6R-3'-UTR group was dramatically lower than that of the control mimic + IL-6R-3'-UTR group (P<0.001); the hsa-miR-21-5p + IL-6R-3'-UTR-mutant group showed no significant change compared with the control mimic + IL-6R-3'-UTR-mutant group (P>0.05) (Figure 6B). Therefore, the luciferase assay results implied that hsa-miR-21-5p could directly bind to the IL-6R-3'-UTR and suppress IL-6R expression. Next, we explored how the hsa-miR-21-5p regulates IL-6R in HMEC-1 cells. We transfected the control mimic, hsa-miR-21-5p mimic, and hsa-miR-21-5p inhibitor into HMEC-1 cells and detected the IL-6R mRNA and protein levels after 48 hours of transfection. The qRT-PCR results showed that the hsa-miR-21-5p expression levels were elevated in the hsa-MiR-21-5p mimic group (P<0.001) and decreased in the hsa-miR-21-5p inhibitor group (P<0.01) (Figure 6C). The IL-6R mRNA levels were down-regulated in the hsa-miR-21-5p mimic group (P<0.001) and up-regulated in the miR-21-5p inhibitor group (P<0.001) (Figure 6D). Similarly, compared to the control mimic group, the IL-6R proteins were decreased in the hsa-miR-21-5p mimic group, and increased in the hsa-miR-21-5p mimic (Figure 6E,6F). Thus, these results suggest that hsa-miR-21-5p could bind the 3'-UTR of IL-6R and suppress IL-6R expression.
Overexpressed hsa-miR-21-5p alleviates cellular injury in HMEC-1
According to the above data, we hypothesized that hsa-miR-21-5p could reduce the damage of OGD treatment by suppressing IL-6R expression. To validate this hypothesis, we transfected control mimic, hsa-miR-21-5p mimic, and hsa-miR-21-5p inhibitor into HMEC-1 cells respectively, and the 6-hour OGD-treated cells, then quantify the cell viabilities and apoptosis levels in different groups. In non-OGD treated cells, overexpressed hsa-miR-21-5p mimic was shown to mildly increase cell viability (P<0.05) and decrease apoptosis level (P<0.05) (Figure 7A,7B). Decreased hsa-miR-21-5p exerted no significant changes on cell viability (P>0.05) and apoptosis level (P>0.05) compared to the control mimic group. The overexpressed hsa-miR-21-5p in OGD-treated HMEC-1 cells, cell viability, and apoptosis levels showed a significant elevation (P<0.05) and reduction (P<0.05), respectively. Reduced hsa-miR-21-5p could induce dramatically descending cell viability (P<0.01) and increased apoptosis (P<0.01) (Figure 7C,7D). Hence, hsa-miR-21-5p could alleviate the damage induced by OGD treatment in HMEC-1 cells.
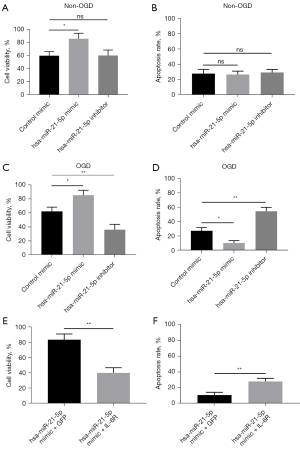
Then, we co-transfected the hsa-miR-21-5p mimic and green fluorescent protein (GFP) plasmids or hsa-miR-21-5p mimic and IL-6R plasmids into HMEC-1 cells. After 48 hours of transfection, we observed that the cell viabilities were decreased and the rates of apoptosis were elevated in hsa-miR-21-5p mimic + IL-6R group compared to hsa-miR-21-5p mimic + GFP group cells (Figure 7E,7F). Therefore, overexpressed IL-6R could reduce the protection function of hsa-miR-21-5p in OGD-treated HMEC-1 cells.
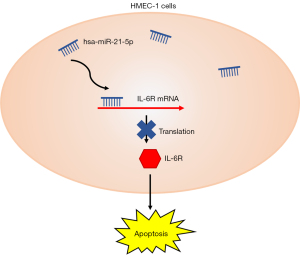
In general, the hsa-miR-21-5p was significantly declined in HMEC-1 after OGD treatment. Overexpression of hsa-miR-21-5p could enhance cell viability and reduce apoptosis by suppressing IL-6R expression in the OGD environment (Figure 8).
Discussion
In this study, we found that the expression of hsa-miR-21-5p was significantly changed in the GSE158313 datasets and exhibited a decreased expression in the peripheral blood of clinical ischemic stroke patients compared to healthy people. Multiple recent publications have reported the important role of hsa-miR-21-5p in the ischemic stroke process. Dong et al. reported that miR-21 and miR-24 harbor potential as diagnostic predictors in ischemic stroke patients (21). Zhang et al. also reported that the miR-21 levels were changed with the progression of ischemic stroke disease in 2007 (22). Thus, we suggest that hsa-miR-21-5p could be a clinical biomarker in future ischemic stroke diagnoses.
Interestingly, several articles have reported different and conflicting observations of the variation of hsa-miR-21-5p in peripheral blood. In stroke and atherosclerosis patients, the expression of miR-21 has been reported to be significantly elevated in peripheral blood samples compared to the control group (21). In male rats that has been subjected to middle cerebral artery occlusion (MCAO) surgery, the miR-21 has also been shown to be up-regulated in ischemic brain tissues compared to sham rats (22). Similarly, an in-situ hybridization assay showed that miR-21 was increased in neurons around the ischemic boundary region, and the cultured primary neurons separated from the ischemic boundary also showed increased miR-21 expression compared to sham region primary neurons (23). However, other articles have suggested that miR-21 is reduced in cultured cortical primary neurons after OGD treatment and also presents decreased expression in the brain tissues of the central MCAO region (24). In this study, we explored the hsa-miR-21-5p levels in the peripheral blood of human ischemic stroke patients, and both the bioinformatics analysis and qRT-PCR results showed that hsa-miR-21-5p was reduced in patients 2 days after stroke compared to controls. Meanwhile, hsa-miR-21-5p was down-regulated in the OGD-treated HMEC-1 cells. We speculate that the conflicting miR-21 expression variation in stroke patients or animal and cell models is caused by the different regions of samples, the difference in sampling time point after stroke, and the disparate cell lines in studies. Therefore, future miR-21-5p and stroke-related studies should pay attention to the sampling time and sample types, which may cause variation from previous articles.
The protective function of hsa-miR-21-5p in the ischemic stroke process is well accepted in the field, but the mechanism by which hsa-miR-21-5p protects cells is complex and still unclear. A study suggested that miR-21 could up-regulate the PI3K/Akt signaling pathway by suppressing PTEN expression, then inhibit caspase-3 levels to reduce the cell apoptosis ratio in OGD-treated cells or MCAO-treated mice (25). Also, miR-21 could inhibit p53 expression to down-regulate apoptosis-related protein Bcl-2 and Bax levels after OGD/reperfusion (R) treatment, then alleviate cellular apoptosis and impairment (24). Besides, in the OGD-treated newborn rat brain, miR021 could bind CCL3 mRNA and inhibit its expression to suppress the phosphorylation of p65, then inhibit the NF-κB signaling pathway to reduce the inflammatory reaction (26). In this study, we explored the role of hsa-miR-21-5p in microvascular epithelial cells in the ischemia-reperfusion process, the luciferase reporter assay results showed that hsa-miR-21-5p could bind the 3'-UTR of IL-6R mRNA then regulate apoptosis-related proteins including Bax, Bcl-2, cleaved-caspase-3, and cleaved-caspase-9, further suppressing cellular apoptosis and enhancing cell viability.
IL-6R also named CD126, as a cytokine receptor of IL-6, could regulate multiple biological process, such as cell proliferation, cell differentiation, and immunization. Several studies have reported that the serum IL-6 concentration shows a positive relationship with impaired volumes of stroke patients in magnetic resonance imaging (MRI) images (P<0.001, r=0.7) (27). In the brain of stroke patients, IL-6 could activate the serum amyloid A (SAA) pathway and induce neutrophil cells to invade into ischemic neurons, and damage cells (28,29). Some researchers suggest that the damaging effects of IL-6R are caused by excessive inflammation and blood coagulation, which then influence the JAK-STAT pathways to upregulate STAT3 expression, resulting in elevated inflammation to induce cell death, and injection of the IL-6R antibody after MCAO increased cell death and infarct volume (30-33). In this study, we found that the level of IL-6R in OGD treated cells was up-regulated, which may affect the IL-6 related signaling pathway and lead to the increase of apoptosis related proteins.
In general, this study focused on the in vivo ischemic stroke models of human microvascular epithelial cells and firstly reports that hsa-miR-21-5p could directly bind to IL-6R in HMEC-1 cells. Further, hsa-miR-21-5p could suppress IL-6R expression to decrease apoptosis levels and enhance cell viability. The results of this study provide a new sight into the injury of microvascular epithelial cells in cerebral ischemia and present a new potential target for future stroke treatment.
Conclusions
Hsa-miR-21-5p could bind to the IL-6R gene and suppress IL-6R expression, thus alleviating the damage of OGD treatment in HMEC-1 cells.
Acknowledgments
Funding: This work was supported by the Qiqihar Academy of Medical Sciences Clinical Research Project (No. QMSI2020L-04).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6451/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6451/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6451/coif). All authors report that this work was supported by the Qiqihar Academy of Medical Sciences Clinical Research Project (No. QMSI2020L-04). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The Second Affiliated Hospital of Qiqihar Medical University (No. 2021-0507), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Saini V, Guada L, Yavagal DR. Global Epidemiology of Stroke and Access to Acute Ischemic Stroke Interventions. Neurology 2021;97:S6-16. [Crossref] [PubMed]
- Martinez B, Peplow PV. Immunomodulators and microRNAs as neurorestorative therapy for ischemic stroke. Neural Regen Res 2017;12:865-74. [Crossref] [PubMed]
- Balami JS, Chen RL, Buchan AM. Stroke syndromes and clinical management. QJM 2013;106:607-15. [Crossref] [PubMed]
- Panagal M, Biruntha M, Vidhyavathi RM, et al. Dissecting the role of miR-21 in different types of stroke. Gene 2019;681:69-72. [Crossref] [PubMed]
- Krishnan R, Mani P, Sivakumar P, et al. Expression and methylation of circulating microRNA-510 in essential hypertension. Hypertens Res 2017;40:361-3. [Crossref] [PubMed]
- Sekar D, Venugopal B, Sekar P, et al. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016;582:14-8. [Crossref] [PubMed]
- Zhou C, Zhao X, Duan S. The role of miR-543 in human cancerous and noncancerous diseases. J Cell Physiol 2021;236:15-26. [Crossref] [PubMed]
- Liu W, Chen X, Zhang Y. Effects of microRNA-21 and microRNA-24 inhibitors on neuronal apoptosis in ischemic stroke. Am J Transl Res 2016;8:3179-87.
- Zhang J, Luo CJ, Xiong XQ, et al. MiR-21-5p-expressing bone marrow mesenchymal stem cells alleviate myocardial ischemia/reperfusion injury by regulating the circRNA_0031672/MiR-21-5p/programmed cell death protein 4 pathway. J Geriatr Cardiol 2021;18:1029-43. [Crossref] [PubMed]
- Li JW, Wei L, Han Z, et al. Mesenchymal stromal cells-derived exosomes alleviate ischemia/reperfusion injury in mouse lung by transporting anti-apoptotic MiR-21-5p. Eur J Pharmacol 2019;852:68-76. [Crossref] [PubMed]
- Surina S, Fontanella RA, Scisciola L, et al. miR-21 in Human Cardiomyopathies. Front Cardiovasc Med 2021;8:767064. [Crossref] [PubMed]
- Nasci VL, Chuppa S, Griswold L, et al. MiR-21-5p regulates mitochondrial respiration and lipid content in H9C2 cells. Am J Physiol Heart Circ Physiol 2019;316:H710-21. [Crossref] [PubMed]
- Cao LQ, Yang XW, Chen YB, et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer 2019;18:148. [Crossref] [PubMed]
- Zhang J, Li D, Zhang R, et al. The miR-21 potential of serving as a biomarker for liver diseases in clinical practice. Biochem Soc Trans 2020;48:2295-305. [Crossref] [PubMed]
- Hurst RT, Lee RW. Increased incidence of coronary atherosclerosis in type 2 diabetes mellitus: mechanisms and management. Ann Intern Med 2003;139:824-34. [Crossref] [PubMed]
- Sekar D, Shilpa BR, Das AJ. Relevance of microRNA 21 in Different Types of Hypertension. Curr Hypertens Rep 2017;19:57. [Crossref] [PubMed]
- Winek K, Lobentanzer S, Nadorp B, et al. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc Natl Acad Sci U S A 2020;117:32606-16. [Crossref] [PubMed]
- Krug T, Gabriel JP, Taipa R, et al. TTC7B emerges as a novel risk factor for ischemic stroke through the convergence of several genome-wide approaches. J Cereb Blood Flow Metab 2012;32:1061-72. [Crossref] [PubMed]
- O'Connell GC, Petrone AB, Treadway MB, et al. Machine-learning approach identifies a pattern of gene expression in peripheral blood that can accurately detect ischaemic stroke. NPJ Genom Med 2016;1:16038. [Crossref] [PubMed]
- Witkowski M, Witkowski M, Saffarzadeh M, et al. Vascular miR-181b controls tissue factor-dependent thrombogenicity and inflammation in type 2 diabetes. Cardiovasc Diabetol 2020;19:20. [Crossref] [PubMed]
- Dong YF, Guo RB, Ji J, et al. S1PR3 is essential for phosphorylated fingolimod to protect astrocytes against oxygen-glucose deprivation-induced neuroinflammation via inhibiting TLR2/4-NFκB signalling. J Cell Mol Med 2018;22:3159-66. [Crossref] [PubMed]
- Zhang K, Zhao Z, Yu J, et al. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem 2018;119:6045-56. [Crossref] [PubMed]
- Zhang L, He X, Jin T, et al. Long non-coding RNA DLX6-AS1 aggravates hepatocellular carcinoma carcinogenesis by modulating miR-203a/MMP-2 pathway. Biomed Pharmacother 2017;96:884-91. [Crossref] [PubMed]
- Xu XM, Qian JC, Deng ZL, et al. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett 2012;4:339-45. [Crossref] [PubMed]
- Luo X, Zhang X, Zhao T, et al. A preliminary study on the proinflammatory mechanisms of Treponema pallidum outer membrane protein Tp92 in human macrophages and HMEC-1 cells. Microb Pathog 2017;110:176-83. [Crossref] [PubMed]
- Mao B, Xiao H, Zhang Z, et al. MicroRNA-21 regulates the expression of BTG2 in HepG2 liver cancer cells. Mol Med Rep 2015;12:4917-24. [Crossref] [PubMed]
- Tsai PC, Liao YC, Wang YS, et al. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J Vasc Res 2013;50:346-54. [Crossref] [PubMed]
- Liu FJ, Lim KY, Kaur P, et al. microRNAs Involved in Regulating Spontaneous Recovery in Embolic Stroke Model. PLoS One 2013;8:e66393. [Crossref] [PubMed]
- Buller B, Liu X, Wang X, et al. MicroRNA-21 protects neurons from ischemic death. FEBS J 2010;277:4299-307. [Crossref] [PubMed]
- Yan H, Huang W, Rao J, et al. miR-21 regulates ischemic neuronal injury via the p53/Bcl-2/Bax signaling pathway. Aging (Albany NY) 2021;13:22242-55. [Crossref] [PubMed]
- Yang Q, Yang K, Li A. microRNA-21 protects against ischemia-reperfusion and hypoxia-reperfusion-induced cardiocyte apoptosis via the phosphatase and tensin homolog/Akt-dependent mechanism. Mol Med Rep 2014;9:2213-20. [Crossref] [PubMed]
- Liu J, Zhang S, Huang Y, et al. miR-21 protects neonatal rats from hypoxic-ischemic brain damage by targeting CCL3. Apoptosis 2020;25:275-89. [Crossref] [PubMed]
- Matsuda S, Wen TC, Morita F, et al. Interleukin-6 prevents ischemia-induced learning disability and neuronal and synaptic loss in gerbils. Neurosci Lett 1996;204:109-12. [Crossref] [PubMed]
(English Language Editor: J. Jones)


