
XTP1 facilitates the growth and development of gastric cancer by activating CDK6
Highlight box
Key findings
• XTP1 can promote the development and progression of GC cells by activating CDK6.
What is known and what is new?
• XTP1 is overexpressed in tumor tissues and regulates cancer progression.
• XTP1 is highly expressed in tumor tissues of GC and related to GC growth and development via promoting the expression of CDK6.
What is the implication, and what should change now?
• The XTP1-CDK6 axis may be a potential therapeutic target for GC, and it can be used in further studies on GC treatment.
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the fourth leading cause of cancer death globally (1). In China, nearly 460,000 new cases of GC are diagnosed annually, and more than 350,000 GC-related deaths occur each year (2). Widespread tumor migration and invasion contribute to its high mortality rate. Despite the progress in treatment strategies for GC, the efficacy of conventional therapies, including surgery, chemotherapy, and radiotherapy, is still unsatisfactory. Major advances in targeted therapy and immunotherapy have started to alter the clinical landscape of GC treatment and prognosis. Moreover, numerous stimulatory and inhibitory factors have been proven to play essential roles in the growth and metastasis of GC (3). However, it is still necessary to further investigate the underlying mechanism in the pathogenesis of GC to identify useful therapeutic targets.
Hepatitis B virus X protein (XTP1), also known as BRCA1/BRCA2-containing complex, subunit 3 (BRCC3) and DEP domain containing 1B (DEPDC1B), was a gene screened by suppression subtractive hybridization in our study on the transactivation of XTP1. XTP1 is involved in the promotion of myoblast proliferation and the retraining of precocious myogenic differentiation in skeletal myogenesis (4). Notably, accumulating evidence has revealed the protumor effects of XTP1 in several tumors, including breast cancer (5), non-small cell lung cancer (6), bladder cancer (7,8), prostate cancer (9), malignant melanoma (10), hepatocellular carcinoma (11), pancreatic cancer (12,13), and so on (14,15). Several signaling pathways, including nuclear factor kappa-B (NF-κB) signaling (8), Wingless (Wnt)/β-catenin signaling (6), protein kinase B (Akt)/glycogen synthase kinase 3 beta (GSK3β)/Snail signaling (13), ras-related C3 botulinum toxin substrate 1 (Rac1)-extracellular regulated protein kinases 1/2 (ERK1/2) signaling (15), Rac1-p21 protein activated kinase 1 (PAK1) signaling (9), and PAK1-LIM domain kinase 1 (LIMK1)-cofilin1 signaling (12), are involved in tumorigenesis and malignant progression regulated by XTP1. Nevertheless, the role and intrinsic molecular mechanism of XTP1 in gastrointestinal cancer, especially GC, have not been confirmed or clarified.
In the present study, we intended to confirm the effects of XTP1 on GC progression and investigate the mechanisms through which XTP1 regulates tumor development. We present the following article in accordance with the ARRIVE and MDAR reporting checklists (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5933/rc).
Methods
Antibodies
The following antibodies were used in this study: anti-XTP1 (Abcam, USA, ab124182, 1:1,000), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Bioworld, USA, AP0063, 1:3,000), anti-rabbit immunoglobulin (IgG) (Beyotime, China, A0208, 1:3,000), anti-Ki-67 (Abcam, USA, Ab16667, 1:200), anti-rabbit IgG (Abcam, USA, ab6721, 1:400), anti-cluster of differentiation (CD)44 (Abcam, USA, ab157107, 1:2000), anti-cyclin-dependent kinase 6 (CDK6) (Abcam, USA, ab151247, 1:1,000), anti-ras homolog family member U (RHOU) (Abcam, USA, ab80315, 1:1,000), and anti-rabbit IgG (Beyotime, China, A0208, 1:3,000).
Cell culture
Human GC cell lines (AGS, SGC-7901, BGC-823, and MGC-803) (obtained from the Cell Bank of the Chinese Academy of Sciences) were cultured in Roswell Park Memorial Institute (RPMI) 1640 (Corning, USA, 10-040-CVB) with 10% fetal bovine serum (FBS) (Invitrogen, USA, 16000-044) at 37 ℃ and 5% CO2.
XTP1 expression analysis using the Gene Expression Profiling Interactive Analysis (GEPIA) database
The expression level of XTP1 in tumor tissue and adjacent normal tissue of GC was investigated using the GEPIA database (http://gepia.cancer-pku.cn/index.html).
Patients and pathological tissue specimens
Pathological tissue specimens for immunohistochemistry (IHC) assays were collected from GC patients who underwent surgical resection at the First Affiliated Hospital of Xi’an Jiaotong University from December 2013 to January 2017. A total of 196 GC samples (including 22 adjacent normal tissue samples) were assessed in this study. No patients received any antitumor therapy before surgical resection. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the clinical ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF2020LSK-103). The study was a retrospective study, and written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements, so it was waived by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
IHC assay
Tissue specimens were embedded in paraffin, sliced into serial 4-nm sections, deparaffinized, and rehydrated. The slides were incubated with primary antibodies overnight at 4 ℃. Subsequently, secondary antibodies were added and incubated for 60 min at 24 ℃. The slides were stained with hematoxylin (Baso, China, BA4041), dehydrated and cover-slipped. Images of the stained slides were obtained using an optical microscope. All of the stained slides were independently evaluated and scored by two pathologists, and an agreement was reached after careful discussion if discrepancies occurred.
Lentivirus construction and cell transfection
The target gene was used as a template and evaluated using design software (http://tools.genome-engineering.org) to find the interference target sequence. The target sequence of XTP1 was 5'-GCTGCTAGATTGGTAACGTTT-3', and the target sequence of CDK6 was 5'-GCCCAACCAATTGAGAAGTTT-3'. The negative control sequence was 5'-TCTCCGAACGTGTCACGT-3'. Next, lentivirus construction and cell transfection were conducted according to a published protocol (16). The transfected cells with puromycin resistance was screened by culture medium with puromycin (Clontech, USA, 631305).
Western blotting
RIPA lysis buffer was used to lyse the cells. The lysates were washed with phosphate buffered saline (PBS) and centrifuged. A BCA Protein Assay Kit (HyClone, USA, Pierce 23225) was used to detect the abundance of the proteins. The samples were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels for electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes. Then, the antibodies were added and incubated with the membranes. An enhanced chemiluminescence (ECL)-Pus/Kit (Amersham, USA, RPN2232) was used to detect the protein bands.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
TRIzol reagent (Sigma, USA, T9424) was used to extract the total RNA from GC. The complementary DNA (cDNA) was reverse transcribed according to the manufacturer’s instructions (+gDNA WIPER, Vazyme, China, R123-01). Subsequently, RT-qPCR was performed to quantify the RNA expression. Table S1 shows the primer sequences used in this study. The relative gene expression levels were calculated by the 2−ΔΔCt method.
Methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay and Celigo cell counting
The cells were seeded into 96-well plates (1,500 cells/well). Then, the MTT and Celigo cell counting assays were performed according to the published protocols (16).
Colony formation assay
The cells were seeded into six-well plates (500 cells/well). The cells were cultured for 13 days and then washed with PBS. Next, 4% paraformaldehyde (1 mL) was used to fix the cells for 30–60 min. Subsequently, Giemsa dye (500 µL) was used to stain the cells for 10–20 min. Double-distilled water (ddH2O) was then used to wash the cells several times and dry them at 24 ℃. The cell colonies were scanned using a microscope.
Apoptosis assay
Trypsin was used to digest the cells, and the digested cells were centrifuged and fixed with 70% ethanol. The cells were then stained with annexin-V-allophycocyanin (APC). Thereafter, flow cytometry was performed to identify the apoptotic cells. An apoptosis detection kit (eBioscience, USA, 88-8007) was used in this experiment.
Human apoptosis antibody array
The human apoptosis antibody array (Abcam, USA, ab134001) was used in this study according to the manufacturer’s instructions. Briefly, total proteins from the cells were extracted. Biotin-labeled anti-cytokines, horseradish peroxidase-labeled streptavidin, and chemiluminescence detection reagents were added and reacted. Then, the spots on the array were scanned, and the expression levels of the proteins related to apoptosis were analyzed quantitatively.
Cell cycle analysis
The cells were centrifuged and washed with precooled Dulbecco’s phosphate buffered saline (DPBS), and then centrifuged and fixed with precooled 70% ethanol. A 40× propidium iodide (PI) solution (2 mg/mL) was used to dye the cells. Flow cytometry was performed to analyze the cell cycle.
Scratch wound-healing assay
The transfected cells were seeded in 96-well plates (50,000 cells/well). Twenty-four hours later, a 96 Wounding Replicator (VP scientific, USA, VP408FH) was used to draw the scratches across the plate. Next, 2% PBS was added to wash the cells several times. Images were scanned at 0, 24, and 48 h using the Celigo image cytometry system (Nexcelom, USA), and the migration rate was calculated according to the published equation (16).
Transwell migration assay
The cells were seeded in 24-well plates (80,000 cells/well) and diluted using a medium with 30% FBS (100 µL), while a medium with 30% FBS (600 µL) was added to the basolateral chamber. Subsequently, the chambers containing cells were transferred to the basolateral chamber and incubated for 24 h. The non-migrated cells were swabbed, and a 4% paraformaldehyde and Giemsa dye were then added to fix and stain the cells, respectively. A microscope was used to count the cells at 100× magnification.
Xenograft tumor model
A protocol was prepared before the study without registration. A total of 20 4-week-old (weight about 20 g) female Balb/c nude mice were obtained from Shanghai Lingchang Biology (China). The mice were randomly divided into short hairpin (sh)Ctrl and shXTP1 groups via random number table method, with 10 mice in each group. Next, the mice were subcutaneously injected with transfected SGC-7901 cells (4×106) at the right forelimb. Tumor data collection began after 10 days. Bioluminescence imaging was performed before the mice were sacrificed, and the mice were sacrificed 3 weeks later. Then, the tumor volumes and weights were measured. Hematoxylin and eosin (HE) staining and IHC assays were further performed for the subcutaneous tumor tissues. Animal experiments were performed under a project license (No. XJTULAC2021-1341) granted by the Institutional Animal Care Committee of the Animal Experimental Center of Xi’an Jiaotong University, in compliance with the institutional guidelines for the care and use of animals.
Human GeneChip assay
The Human GeneChip assay was performed with the 3’ IVT Plus Kit (Affymetrix, USA). The double-stranded cDNA of the target gene was transcribed to synthesize complementary RNA (cRNA). The cRNA was then purified, quantified, and labeled. The Pre-Hybridization Mix was added and reacted with the GeneChip. Next, the Hybridization Master Mix was mixed with the labeled cRNA, and the mixture was added and reacted with the GeneChip. A GeneChip Scanner and software were used to scan and analyze the GeneChip, respectively. The raw data were filtered and standardized for further bioinformatics analysis, which was performed using Ingenuity Pathway Analysis (IPA) software.
Statistical analysis
GraphPad Prism 8.3.0 was used to analyze the data. The data were expressed as the mean ± standard error of the mean. T-tests, one-way analysis of variance (ANOVA), and two-way repeated-measures ANOVA were used to compare the differences. A P value <0.05 indicated that there was a statistically significant difference between the two groups.
Results
XTP1 was overexpressed in GC tissues
To investigate the effects of XTP1 on GC, we evaluated the XTP1 expression level in tumor tissues and paracarcinoma tissues of GC patients by IHC. According to the IHC staining results, the XTP1 protein was significantly overexpressed in tumor tissues compared with adjacent normal tissues (Figure 1A,1B). Furthermore, Spearman rank correlation analysis showed that the expression of XTP1 was obviously related to the pathological grade and negatively related to patient age (Table 1). The GEPIA database analysis also suggested that XTP1 was overexpressed in the tumor tissues of GC patients (Figure 1C). In addition, RT-qPCR showed that XTP1 was highly expressed in GC cell lines, especially BGC-823 and SGC-7901 (Figure 1D). All of the above results indicated that XTP1 was overexpressed in GC tumor tissues.
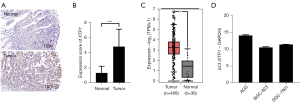
Table 1
| Characteristic | Number | XTP1 expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.008 | |||
| ≤56 | 99 | 52 | 47 | |
| >56 | 97 | 69 | 28 | |
| Sex | 0.671 | |||
| Male | 135 | 82 | 53 | |
| Female | 61 | 39 | 22 | |
| Pathological grade | 0.000 | |||
| I/II | 69 | 60 | 9 | |
| III | 106 | 42 | 64 | |
| NA | 21 | |||
| T stage | 0.972 | |||
| T1 | 4 | 3 | 1 | |
| T2 | 25 | 13 | 12 | |
| T3 | 116 | 65 | 51 | |
| T4 | 11 | 6 | 5 | |
| NA | 40 | |||
| Lymph node metastasis | 0.125 | |||
| N0 | 104 | 63 | 41 | |
| N1 | 34 | 14 | 20 | |
| N2 | 12 | 8 | 4 | |
| N3 | 6 | 2 | 4 | |
| NA | 40 | |||
| Stage | 0.076 | |||
| I | 24 | 15 | 9 | |
| II | 79 | 48 | 31 | |
| III | 39 | 18 | 21 | |
| IV | 14 | 6 | 8 | |
| NA | 40 | |||
| Total | 196 | 121 | 75 | – |
GC, gastric cancer; NA, not applicable.
XTP1 depletion suppressed GC development and progression in vitro
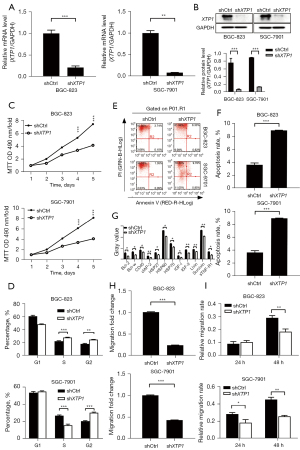
To further explore the role of XTP1 in GC development, a lentivirus containing shRNA against XTP1 was constructed and transfected into the BGC-823 and SGC-7901 cell lines to downregulate the expression of XTP1. RT-qPCR and Western blotting showed that the expression of XTP1 was inhibited after transfection (Figure 2A,2B). Also, MTT and cell cycle assays showed that XTP1 depletion restrained the proliferation of GC cells (Figure 2C,2D). Subsequently, the effect of XTP1 knockdown on the apoptosis of GC cells was investigated by flow cytometry and the human apoptosis antibody array. The apoptosis of GC cell lines was significantly promoted (Figure 2E,2F). Additionally, the expression levels of proteins related to cell apoptosis [including B-cell CLL/lymphoma 2 (Bcl-2), BCL2-like 2 (Bcl-w), CD40, clAP-2, heat shock protein (HSP)27, HSP60, HSP70, insulin-like growth factors (IGF)-I, IGF-II, Livin, Survivin, and soluble tumor necrosis factor receptor 1 (sTNF-R1)] were reduced (Figure 2G) when XTP1 was downregulated. Furthermore, inhibition of the migratory ability was observed in transfected GC cells (Figure 2H,2I). Taken together, these findings indicated that the downregulation of XTP1 significantly inhibited the proliferation and migration, and promoted the apoptosis of GC cells.
XTP1 depletion inhibited GC growth in vivo
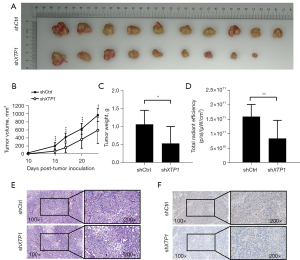
Given the effects of XTP1 knockdown on GC cells in vitro, we further evaluated its role in the growth of GC in vivo. The tumor volumes and weights were significantly decreased after XTP1 depletion (Figure 3A-3C). In vivo imaging also showed an obvious reduction in the total fluorescence intensity after XTP1 depletion (Figure 3D). Notably, we performed HE staining and IHC assays to evaluate the Ki-67 expression in the mice tumor tissues. A decrease in the Ki-67 expression level was observed in mice that were subcutaneously injected with XTP1-depleted cells (Figure 3E,3F). Collectively, the knockdown of XTP1 obviously restrained the growth of GC in vivo.
XTP1 overexpression facilitated GC development and progression in vitro
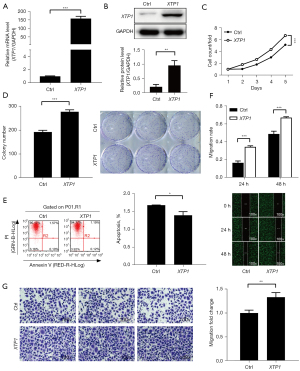
From the above experiments, we found that XTP1 depletion inhibited GC growth and development. However, whether the overexpression of XTP1 could also impact GC development was unclear. Thus, we constructed lentivirus and transfected SGC-7901 cells to upregulate XTP1 expression. The results of RT-qPCR and Western blotting showed that XTP1 was overexpressed following transfection (Figure 4A,4B). The Celigo cell counting and colony formation assays suggested that XTP1 overexpression could promote GC cell proliferation (Figure 4C,4D). In addition, flow cytometry indicated that XTP1 overexpression was inhibited in SGC-7901 cells (Figure 4E). Additionally, the Transwell migration assays and scratch wound-healing assays showed that XTP1 upregulation promoted the migration of SGC-7901 cells (Figure 4F,4G). Taken together, the above findings suggested that XTP1 overexpression significantly facilitates the growth of GC cells and suppresses GC cell apoptosis.
Identification of downstream molecules of XTP1 in GC cells
To elucidate the mechanism underlying the effects of XTP1 on GC cells, we conducted a human GeneChip assay and bioinformatics analysis. Our GeneChip assay identified 642 differentially expressed genes (DEGs) in the SGC-7901 cells with versus without XTP1 depletion (Figure 5A-5C). We also identified 407 genes that were upregulated and 235 genes that were downregulated after XTP1 knockdown by hierarchical clustering of the DEGs (Figure 5C). Further IPA highlighted the enrichment of 642 DEGs related to cell death and survival (Figure 5D), as well as the inhibition of signaling pathways, including relaxin signaling, small cell lung cancer signaling, and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling (Figure 5E).
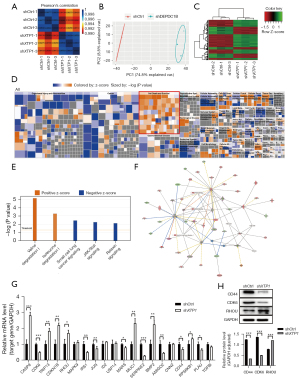
Moreover, the gene interaction network based on IPA showed that XPT1 could indirectly impact the downstream molecules involved in relaxin signaling, small cell lung cancer signaling, and JAK/STAT signaling, including AKT2, BCL2-like 1 (BCL2L1), caspase 9 (CASP9), CDK6, cyclin-dependent kinase inhibitor 1B (CDKN1B), cyclin-dependent kinase inhibitor 2B (CDKN2B), insulin receptor substrate 1 (IRS1), Jun proto-oncogene (JUN), mitogen-activated protein kinase 1 (MAPK1), muscle RAS oncogene homolog (MRAS), and so on (Figure 5F). These molecules were selected for further characterization. RT-qPCR showed that XTP1 knockdown induced the upregulation of seven genes [CASP9, thyroid hormone receptor interactor 13 (TRIP13), CDKN1B, RHOU, mucin 1 (MUC1), bone morphogenetic protein 2 (BMP2), and ribosomal protein S6 kinase (RPS6KB1)] and the downregulation of eight genes [CDK6, IRS1, JUN, methionyl-tRNA synthetase (MARS), serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2), adhesion molecule with Ig like domain 2 (AMGO2), CD44, and plasminogen activator, urokinase (PLAU)] (Figure 5G).
Previous studies have suggested that CD44 is associated with the tumorigenicity, invasion, and lymph node metastasis of cancer cells (17,18), while CDK6 and RHOU are associated with the cell cycle and cell migration, respectively (19-21). Thus, we conducted a Western blot analysis to confirm the expressions of CD44, CDK6, and RHOU. The results showed that the knockdown of XTP1 induced RHOU upregulation and the downregulation of CDK6 and CD44 (Figure 5H). In summary, XTP1 might affect the biological functions of GC cells via CDK6, CD44, and RHOU.
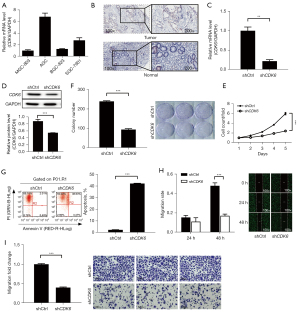
Knockdown of CDK6 inhibited the growth of GC cells
CDK6 inhibitors have been used in the development of antitumor drugs (20,22). Therefore, we selected CDK6 for further investigation. First, we confirmed the expression level of CDK6 in GCs and found that CDK6 was overexpressed in the GC cell lines (Figure 6A) and tumor tissues (Figure 6B) by RT-qPCR and IHC analysis, respectively. Next, we constructed lentiviruses containing shRNA targeting CDK6 and transfected SGC-7901 cells. CDK6 expression was significantly suppressed by lentivirus containing shRNA against CDK6 (Figure 6C,6D). Proliferation assays illustrated the inhibitory effect of CDK6 depletion on GC cell growth (Figure 6E,6F). The promoting effect of CDK6 knockdown on GC cell apoptosis was also observed (Figure 6G). In addition, the migration of GC cells was evaluated by Transwell migration and scratch wound-healing assays. The results indicated that CDK6 knockdown suppressed the migration of GC cells (Figure 6H,6I). Together, the above results highlighted the critical role of CDK6 in GC.
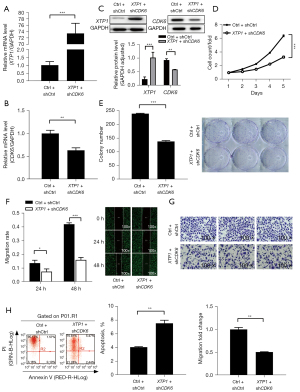
XTP1 depletion restrained GC cell growth and development by inhibiting CDK6
To verify whether XTP1 facilitates the development of GC by regulating CDK6, we cotransfected SGC-7901 cells with lentiviruses for overexpressing XTP1 and silencing CDK6. RT-qPCR and Western blot analysis showed that XTP1 was overexpressed, while CDK6 was downregulated (Figure 7A-7C). We further evaluated GC cell proliferation, apoptosis, and migration. CDK6 depletion completely blocked the XTP1 overexpression-induced enhancement of SGC-7901 cell proliferation (Figure 7D,7E) and migration (Figure 7F,7G). In addition, flow cytometry suggested that the reduction in the apoptosis of GC cells induced by XTP1 overexpression was alleviated by CDK6 knockdown (Figure 7H). Taken together, these findings indicated that CDK6 is a downstream molecule of XTP1. Therefore, XTP1 may facilitate GC cell growth and development by activating CDK6.
Discussion
In this study, the role of XTP1 in the proliferation, apoptosis, and migration of GC cells was identified by depleting or overexpressing XTP1, and the potential molecular mechanism was explored using a human GeneChip assay combined with bioinformatics analysis.
XTP1 was upregulated in GC cells and tumor tissues. Further investigation revealed a positive relationship between high XTP1 expression levels and the malignant degree of GC. Moreover, XTP1 depletion suppressed the growth and migration of GC cells, while GC cell apoptosis was promoted and the expression of apoptosis-related proteins was inhibited by XTP1 depletion. These results revealed the important regulatory effects of XTP1 on GC development. To further verify the effects of XTP1 on GC cell growth, we upregulated XTP1 and found that overexpression could obviously increase the proliferative and migratory potential of GC cells and inhibit GC cell apoptosis. The above results comprehensively confirmed that XTP1 is a potent tumor promoter in GC.
XTP1 was previously recognized as a tumor promoter of bladder cancer by targeting SHC1 (7). Later, Tao et al. found that XTP1 could activate the NF-κB signaling pathway by targeting TRAF2, thereby promoting the development of bladder cancer (8). Similarly, XTP1 was related to cell proliferation and epithelial-mesenchymal transition in prostate cancer via Rac1-PAK1 signaling (9). In addition, XTP1 was reported to activate Wnt/β-catenin signaling to enhance the migration and invasion of non-small cell lung cancer (6). These previous studies indicated that XTP1 plays a role in tumorigenesis as a tumor promoter by targeting downstream molecules. In our study, we conducted a human GeneChip assay combined with bioinformatics analysis to identify the possible downstream molecules of XTP1 in GC and hypothesized that CDK6 is a potential downstream molecule of XTP1. Interestingly, through RT-qPCR and Western blot analysis, we observed the suppression of CDK6 expression induced by XTP1 knockdown. Next, we detected whether CDK6 is involved in GC cell development. We used a lentivirus to silence CDK6 and evaluated the impact of CDK6 knockdown on GC cells. We observed that CDK6 silencing obviously restrained the proliferation and migration, and facilitated the apoptosis of GC cells, indicating that CDK6 affects the growth of GC cells. Thus, we believe that both XTP1 and CDK6 participate in GC tumorigenesis.
CDK6 is a member of the cyclin-dependent kinase family, which can combine with cyclins to form heterodimers and facilitate cell cycle progression (19,23). In the G1/S phase, CDK6 can combine with cyclin D and promote the phosphorylation of the tumor suppressor Rb, thereby inducing the release of E2F transcription factors from Rb-mediated inhibition. Furthermore, the activation of E2F-dependent genes accelerates the progression of the cell cycle from the G1 to the S phase and DNA synthesis (19). Although CDK6 is described as a critical regulator in cell cycle progression, the latest studies have confirmed that it also plays a role in hematopoiesis (24), T-cell development (25), and immune responses (26). Moreover, the phenomenon of CDK6 overexpression has been confirmed in several malignancies, including acute myeloid leukemia (27), breast cancer (28), and melanoma (29). Our study also confirmed the overexpression of CDK6 in GC cells and its important role in GC development. The critical role of CDK6 in promoting tumor occurrence and development makes it an attractive target for drug inhibition (30). In this study, XTP1 knockdown resulted in the downregulation of CDK6, indicating that XTP1 might modulate GC development by regulating CDK6 expression. When XTP1 was overexpressed and CDK6 was depleted simultaneously, facilitation of the growth and migration of GC cells induced by XTP1 was attenuated via CDK6 knockdown. CDK6 knockdown also eliminated the inhibitory effect of XTP1 depletion on GC cells. These results revealed that XTP1 knockdown regulated GC progression by inhibiting CDK6 expression. However, whether XTP1 could bind with CDK6 was not verified in this study. Thus, in the next step, we will confirm the interaction of XTP1 and CDK6.
Conclusions
In summary, our findings indicate that XTP1 might be a tumor promoter facilitating GC progression, and XTP1 is overexpressed in GC tumor tissues and relevant to the malignant degree of GC. Both XTP1 and CDK6 play critical roles in GC cell proliferation, migration, and apoptosis. Notably, XTP1 can regulate the growth of GC cells by acting on CDK6. In conclusion, our study may provide a new understanding of the pathogenesis and mechanism of GC. The XTP1-CDK6 axis could be used in further studies on GC treatment.
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Shaanxi Province (No. 2021JM-263).
Footnote
Reporting Checklist: The authors have completed the ARRIVE and MDAR reporting checklists. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5933/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5933/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5933/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the clinical ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University (No. XJTU1AF2020LSK-103). The study was a retrospective study, and written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements, so it was waived by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University. Animal experiments were performed under a project license (No. XJTULAC2021-1341) granted by the Institutional Animal Care Committee of the Animal Experimental Center of Xi’an Jiaotong University, in compliance with the institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Mousa H, Yuan M, Zhang X, et al. MicroRNA-4316 inhibits gastric cancer proliferation and migration via directly targeting VEGF-A. Cancer Cell Int 2020;20:62. [Crossref] [PubMed]
- Figeac N, Pruller J, Hofer I, et al. DEPDC1B is a key regulator of myoblast proliferation in mouse and man. Cell Prolif 2020;53:e12717. [Crossref] [PubMed]
- Zhang L, Du Y, Xu S, et al. DEPDC1, negatively regulated by miR-26b, facilitates cell proliferation via the up-regulation of FOXM1 expression in TNBC. Cancer Lett 2019;442:242-51. [Crossref] [PubMed]
- Yang Y, Liu L, Cai J, et al. DEPDC1B enhances migration and invasion of non-small cell lung cancer cells via activating Wnt/β-catenin signaling. Biochem Biophys Res Commun 2014;450:899-905. [Crossref] [PubMed]
- Lai CH, Xu K, Zhou J, et al. DEPDC1B is a tumor promotor in development of bladder cancer through targeting SHC1. Cell Death Dis 2020;11:986. [Crossref] [PubMed]
- Tao H, Liao Y, Yan Y, et al. BRCC3 Promotes Tumorigenesis of Bladder Cancer by Activating the NF-κB Signaling Pathway Through Targeting TRAF2. Front Cell Dev Biol 2021;9:720349. [Crossref] [PubMed]
- Li Z, Wang Q, Peng S, et al. The metastatic promoter DEPDC1B induces epithelial-mesenchymal transition and promotes prostate cancer cell proliferation via Rac1-PAK1 signaling. Clin Transl Med 2020;10:e191. [Crossref] [PubMed]
- Xu Y, Sun W, Zheng B, et al. DEPDC1B knockdown inhibits the development of malignant melanoma through suppressing cell proliferation and inducing cell apoptosis. Exp Cell Res 2019;379:48-54. [Crossref] [PubMed]
- Dang XW, Pan Q, Lin ZH, et al. Overexpressed DEPDC1B contributes to the progression of hepatocellular carcinoma by CDK1. Aging (Albany NY) 2021;13:20094-115. [Crossref] [PubMed]
- Zhang S, Shi W, Hu W, et al. DEP Domain-Containing Protein 1B (DEPDC1B) Promotes Migration and Invasion in Pancreatic Cancer Through the Rac1/PAK1-LIMK1-Cofilin1 Signaling Pathway. Onco Targets Ther 2020;13:1481-96. [Crossref] [PubMed]
- Liu X, Li T, Huang X, et al. DEPDC1B promotes migration and invasion in pancreatic ductal adenocarcinoma by activating the Akt/GSK3β/Snail pathway. Oncol Lett 2020;20:146. [Crossref] [PubMed]
- Wang L, Tang L, Xu R, et al. DEPDC1B regulates the progression of human chordoma through UBE2T-mediated ubiquitination of BIRC5. Cell Death Dis 2021;12:753. [Crossref] [PubMed]
- Su YF, Liang CY, Huang CY, et al. A putative novel protein, DEPDC1B, is overexpressed in oral cancer patients, and enhanced anchorage-independent growth in oral cancer cells that is mediated by Rac1 and ERK. J Biomed Sci 2014;21:67. [Crossref] [PubMed]
- Ma R, Zhu K, Yuan D, et al. Downregulation of the FBXO43 gene inhibits tumor growth in human breast cancer by limiting its interaction with PCNA. J Transl Med 2021;19:425. [Crossref] [PubMed]
- Chen C, Zhao S, Karnad A, et al. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018;11:64. [Crossref] [PubMed]
- Naor D, Nedvetzki S, Golan I, et al. CD44 in cancer. Crit Rev Clin Lab Sci 2002;39:527-79. [Crossref] [PubMed]
- Nebenfuehr S, Kollmann K, Sexl V. The role of CDK6 in cancer. Int J Cancer 2020;147:2988-95. [Crossref] [PubMed]
- Tadesse S, Yu M, Kumarasiri M, et al. Targeting CDK6 in cancer: State of the art and new insights. Cell Cycle 2015;14:3220-30. [Crossref] [PubMed]
- Zhang JS, Koenig A, Young C, et al. GRB2 couples RhoU to epidermal growth factor receptor signaling and cell migration. Mol Biol Cell 2011;22:2119-30. [Crossref] [PubMed]
- Wang H, Nicolay BN, Chick JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017;546:426-30. [Crossref] [PubMed]
- Tigan AS, Bellutti F, Kollmann K, et al. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene 2016;35:3083-91. [Crossref] [PubMed]
- Laurenti E, Frelin C, Xie S, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 2015;16:302-13. [Crossref] [PubMed]
- Hu MG, Deshpande A, Enos M, et al. A requirement for cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res 2009;69:810-8. [Crossref] [PubMed]
- Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471-5. [Crossref] [PubMed]
- Schmoellerl J, Barbosa IAM, Eder T, et al. CDK6 is an essential direct target of NUP98 fusion proteins in acute myeloid leukemia. Blood 2020;136:387-400. [Crossref] [PubMed]
- Goel S, Wang Q, Watt AC, et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell 2016;29:255-69. [Crossref] [PubMed]
- Read J, Wadt KA, Hayward NK. Melanoma genetics. J Med Genet 2016;53:1-14. [Crossref] [PubMed]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009;9:153-66. [Crossref] [PubMed]


