
E2F1 inhibits cellular senescence and promotes oxaliplatin resistance in colorectal cancer
Highlight box
Key findings
• E2F1 promotes drug resistance in colorectal cancer by inhibiting cell senescence.
What is known and what is new?
• E2F1 is highly expressed in colorectal cancer.
• E2F1 promotes colorectal cancer development and drug resistance by inhibiting cell senescence.
What is the implication, and what should change now?
• Cell senescence is an important mechanism of drug resistance in colorectal tumors.
• E2F1 regulates cell senescence and is a potential target for anti-tumor drug development.
Introduction
Colorectal cancer (CRC) is one of the most prevalent cancers worldwide and the second largest cause of cancer mortality (1). Age is a risk factor, with approximately 90% of cases occurring in people aged over 50 years. However, its frequency among people aged younger than 50 has recently increased (2). Moreover, Chinese and Japanese people are more prone to CRC (3). Other common risk factors for CRC include obesity, red and processed meat, smoking, and alcohol intake (4). Gut microbiota disturbance is also a recently more widely recognized independent risk factor for cancer development (5,6). Chemotherapy is widely used as one of the primary treatments for CRC; it improves the survival time for patients with advanced cancers (7). However, the efficacy of chemotherapy has been considerably reduced due to the potential of long-term chemotherapy to develop chemoresistance and chemotherapy-resistant individuals.
Senescence is defined as irreversible cell cycle arrest caused by cellular stress (8). Intense oncogenic stress, such as radiation, chemotherapy, carcinogens, and oxidative stress, lead to DNA replication overload or damage, thus causing a senescence-associated phenotype (9-11). Cellular senescence is an intrinsic cellular response that limits cell proliferation and prevents tumor development by inducing telomere shortening and cell cycle arrest under normal physiological conditions (12,13). However, when the stress exceeds the threshold levels in tumor cells, cellular senescence acts as a target for anticancer medicines, triggering the activation of the p16/pRb and Arf/TP53/p21 pathways and DNA damage response (DDR), resulting in premature senescence and cell death (14-16). Cellular senescence affects tumor progression and has a great impact on tumor chemoresistance (17,18). For instance, lung cancer cells can inhibit cellular senescence by activating the PTEN/Akt pathway, thereby inducing cisplatin resistance (19). In contrast, doxorubicin treatment induces cellular senescence in the HCT116 and MCF7 cells, thus enhancing their sensitivity to curcumin, caffeine, and thymoquinone (20). However, little is known regarding the function of cellular senescence in chemotherapy resistance in CRC.
The E2F family plays various roles in controlling the progression of the cell cycle (21). For example, E2F1 is a replication stress suppressor and cell cycle regulator; therefore, it can cause programmed cell death and restrict the unrestrained proliferation of cancer cells (22,23). E2F1 is comprehensively expressed in advanced cancer tissues, such as colorectal, breast, testicular, and liver cancer tissues, and promotes their invasion and metastasis, making it a crucial molecular marker for determining tumor prognosis (24-26). In contrast, the inactivation of E2F1 results in the accumulation of DNA replication mediators’ DDR and tumor protein p53 (TP53)-dependent apoptosis of tumor cells (27-30). Therefore, E2F1 acts as a key target for clinical cancer chemotherapy drug development (31). In the investigations of cellular senescence-related processes, E2F1 is predominantly used as a downstream regulatory target of the TP53/p21 and p16/Rb axes (32,33). However, the active involvement of E2F1 in controlling cellular senescence has not been extensively studied.
Herein, we aimed to find whether an active regulation of cellular senescence by E2F1 is involved and its possible role in CRC chemoresistance. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4054/rc).
Methods
Cell culture
Human CRC cell lines, including HCT116 and LoVo, and colonic mucosa cells NCM460 were cultured with Dulbecco’s Modified Eagle Medium (DMEM; high glucose) medium with 10% fetal bovine serum (FBS). Human prostate cancer PC-3 cells and gastric cancer MGC803 cells were cultured in RPMI1640 medium containing 10% FBS. The cell lines were grown at 37 °C in a humidified atmosphere of 5% CO2.
Creation of oxaliplatin-resistant HCT116 (OXA-HCT116) cells
HCT116 cells were treated with 0.1 mol/L oxaliplatin (MCE, HY-17371). The surviving cells were cultured to 80% saturation and passaged twice for confirming survival. The surviving cell population was similarly treated with oxaliplatin concentrations of 0.5 mol/L, 1.0 mol/L, and 2.0 mol/L. In the in vitro study, no more than 10 generations of OXA-HCT116 cells were created.
Plasmid and transfection
Human E2F1 eukaryotic expression plasmid (Lenti-CV186-E2F1), RNA interference plasmid (Lenti-GV298-sh-E2F1), overexpression control plasmid (Lenti-CON294), knockdown control plasmid (Lenti-GV298-sh-Scb) were purchased from Shanghai GeneChem Technology Company.
When the HCT116 cells grew up to 60% of the six-well plates, and plasmid transfection was then completed. Then, 48 hours later, puromycin screening was performed to obtain E2F1 stable overexpression cell line (E2F1), E2F1 knockdown cell lines (sh-E2F1 #1 and sh-E2F1 #2), and corresponding blank control cell lines (Mock and sh-Scb). These cell lines with stable expression were then used for experiments.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
According to the instructions from the manufacturer, the RNeasy Mini Kit (Qiagen, 74,104) was used to extract total cellular RNA, and cDNA was created by reverse transcription using the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa Bio., 6110A). qRT-PCR was performed with StepOneTM Real-Time PCR Systems, and the Ct values of each sample were calculated. The transcript levels were evaluated using the Delta-Delta Ct method, with ACTIN serving as the reference gene. Table 1 lists the sequences of all primers.
Table 1
| Gene | Primers | Sequence | Product size (bp) |
|---|---|---|---|
| E2F1 | Forward | 5'-AAGAACCGCTGTTGTCCCG-3' | 182 |
| Reverse | 5'-GAGGCCGAAGTGGTAGTCG-3' | ||
| β-actin | Forward | 5'-TGCCCATCTACGAGGGGTATG-3' | 156 |
| Reverse | 5'-TCTCCTTAATGTCACGCACGATTT-3' |
qRT-PCR, quantitative reverse transcription polymerase chain reaction; E2F1, E2F translation factor 1.
Western blot
Proteins were extracted using RIPA lysis buffer (Promega, A1731). Then, 30 µg protein from each sample was electrophoresed on 12% Bis-Tris polyacrylamide gel and blotted onto 0.45 µm polyvinylidene fluoride membranes. Afterward, membranes were blocked with 5% skimmed milk. Blots were incubated for 4 hours at 4 °C with the primary antibody and 1 hour at room temperature with the secondary antibody. Normalized to β-actin. Primary antibodies used included E2F1 rabbit antibody (ABclonal, A2067), CDKN2A/p16 rabbit antibody (ABclonal, A0262), CDKN1A/p21 rabbit antibody (ABclonal, A19094), p53 rabbit antibody (ABclonal, A0263), Phospho-p53-S15 rabbit antibody (ABclonal, AP0083) or β-actin mouse antibody (Proteintech, 66009-1-Ig); secondary antibody used included HRP enzyme-labeled goat anti-rabbit IgG (Servicebio, GB23303) and HRP enzyme-labeled goat anti-mouse IgG (Servicebio, GB23301).
MTT assay
The cells were inoculated in 96-well plates at 5,000 cells per well, and five replicate wells per grouping were set up. Then, the medium was replaced with a medium containing oxaliplatin. After 48 hours, the medium in each well was replaced with 20 µL of 3-(4,5)-dimethylthiahiazo(-z-y1)-3,5-di-phenytetrazoliumromide (MTT) solution and incubated for 4 hours in the incubator. Then, the culture medium in the wells was discarded, and dimethyl sulfoxide was added to each well. The absorbance was immediately measured at 490 nm with the enzyme-linked immunosorbent assay. The average absorbance of each group was calculated afterwards.
EdU staining assay
The proliferation of the cells was assessed using the BeyoClickTM EdU-594 Cell Proliferation Assay Kit (Beyotime, C0078S). Then, cells were planted in a 24-well plate and incubated until the morphology of the cells returned to normal. Next, 2X EdU working solution was added and incubated. 4% paraformaldehyde was then added. After washing, cells were incubated with a permeabilization solution. Then, cells were incubated with the Click reaction solution out of the light. The nucleus was stained with DAPI (4',6-diamidino-2-phenylindole). A fluorescence microscope was used to photograph and record.
Cell migration assay
Three parallel horizontal lines were drawn on the back of the 6-well plate. Cells were equally distributed in the wells; then, scars perpendicular to these black lines were scratched through the cells. Afterwards, cells were washed to remove the suspended cells and replaced with fresh serum-free medium. The cells under the microscope were photographed at 0, 12, 24, 36, and 48 hours. Images were processed using Image J software; we calculated the mean value of intercellular distance.
Tranwell assay
A 24-well plate was added with 500 µL DMEM per well, and the Transwell chambers were coated with 50 µL matrix gel and placed into the wells containing medium. After trypsin digestion, cells were centrifuged at 1,000 rpm, resuspended, and counted. A serum-free cell suspension with a suitable concentration was prepared, the cell suspension was added to each chamber, and three replicates were set up for each group. After discarding the medium, cells were soaked in methanol and then stained with crystal violet. Then, the non-invaded cells in the upper chambers were wiped with cotton swabs. the invaded cells were photographed and counted under an inverted microscope. Each chamber was captured at the same magnification for nine fields of vision and counted using Image J.
Immunofluorescence assay
The cells cultivated on round coverslips were fixed with 4% formaldehyde, blocked with 5% skimmed milk, washed, and incubated overnight at 4 °C with chromobox 3 (HP1-γ) primary antibody (Abcam, ab223535). After washing, the cells were stained at room temperature with Fluor 594 goat anti-rabbit IgG secondary antibody (Abcam, ab150080). The cells were then incubated with DAPI (Sigma, D9542). A Nikon A1Si laser confocal microscope was used to capture the images.
Senescence-associated β-galactosidase staining assay
According to the instructions from the manufacturer, the Senescence Associated-β-Galactosidase Staining Kit (Beyotime, C0602) was used to perform the staining. The cells in 6-well plates were fixed at room temperature using a fixative. The cells were then washed with PBS and treated with β-galactosidase staining working solution overnight at 37 °C. The photographs were taken under a light microscope, and the number of positive cells was matched.
Data availability
Cellular senescence gene sets were obtained from AmiGO 2 (http://amigo.geneontology.org/). ChIP-X, PWMs, and Genome Browser programs were used to screen for potential transcription factors that regulate gene expression. The public datasets for analyzing the correlation between gene expression and survival of patients with cancer were available from the Gene Expression Omnibus (GEO) databases (https://www.ncbi.nlm.nih.gov/geo/, accession numbers GSE24551 and GSE41258). The ChIP-Atlas (https://chip-atlas.org/) database was used to assess the potential target genes of transcription factors, and the top 1000 genes were chosen for the Gene Ontology (GO) enrichment analysis. The Gene Set Enrichment Analysis (GSEA) and GO enrichment analysis was performed using Sangerbox 3.0 (http://vip.sangerbox.com/) (34). The relative expression of E2F1 in cell lines was analyzed by the GEO database (GSM5436026; GSM5436028) and the Cancer Cell Line Encyclopedia (CCLE) database (https://sites.broadinstitute.org/ccle). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The survival curves of patients with CRC were estimated using the Kaplan-Meier method with a log-rank test. The measures were expressed as mean ± SEM (standard error of mean), and differences between groups were analyzed using the Student’s t-test. P<0.05 was considered statistically significant. The error bars in the graphs represented standard deviation. Statistical analyses were performed using GraphPad Prism 8.0 software.
Results
E2F1 is an essential transcription factor suppressing cellular senescence in CRC
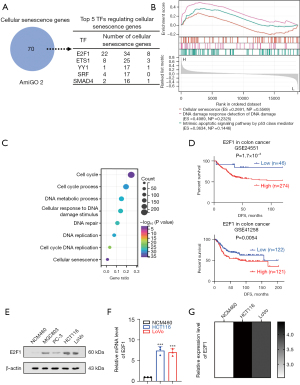
We obtained a cellular senescence-associated gene set from the AmiGO 2 database (http://amigo.geneontology.org/amigo) for investigating the critical transcription factors controlling cellular senescence gene expression. ChIP-X, PWMs, and Genome Browser (35) analyses identified 35 transcription factors affecting these senescence machinery genes. Nine transcription factors were consistently associated with DFS survival in patients with CRC; E2F1 was the highest-ranked transcription factor according to target genes (Figure 1A). The GSEA single gene enrichment analysis of E2F1 revealed that E2F1 was closely related to DNA damage response and TP53-mediated apoptotic pathway, which were required to induce oncogenic stresses for cellular senescence (Figure 1B). Furthermore, the ChIP-Atlas (36,37) (https://chip-atlas.org) database was used to predict the target genes of E2F1; we performed GSEA on the top 1,000 genes. Notably, these genes were enriched for pathways related to the induction of cellular senescence, such as DNA damage and repair (Figure 1C). Moreover, the Kaplan-Meier plots showed that patients with CRC with high E2F1 levels had worse disease-free survival (DFS) than those with low expression (P=1.7×10−3, P=0.0054) (Figure 1D). Moreover, the Western blot and qRT-PCR assays revealed that E2F1 expression was higher in cultured cancer cell lines, particularly CRC cell lines HCT116 and LoVo than in healthy colorectal mucosa (Figure 1E,1F). The analysis results of the CCLE (https://sites.broadinstitute.org/ccle) database and GEO-related sequencing data further corroborated our experimental results (Figure 1G).
E2F1 promotes the proliferation, invasion, and migration of CRC cells
The HCT116 cell line was selected as the next research model to evaluate the importance of E2F1 in colorectal oncogenesis. Furthermore, we established stable cell lines for E2F1 overexpression (E2F1) and E2F1 knockdown (sh-E2F1 #1 and sh-E2F1 #2). Afterward, Western blot and qRT-PCR revealed the expression of E2F1 in the aforementioned cell lines (Figure 2A,2B).
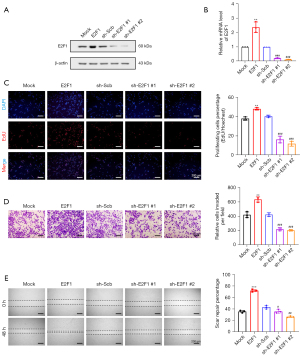
EdU staining was done to examine the proliferative vitality of these CRC cell lines. We noted that the E2F1 overexpression increased the proliferative capacity of these cell lines. Conversely, the E2F1 knockdown reduced it (Figure 2C). The Transwell assay results showed that the E2F1 overexpression increased the invasion capacity of CRC cells, whereas the knockdown of E2F1 decreased it (Figure 2D). Moreover, the scar healing ability of HCT116 overexpressing E2F1 was considerably higher than the control group (Mock); however, opposite results were noted in the knockdown E2F1 group (Figure 2E). These results demonstrated that E2F1, as a pro-oncogene, still promoted proliferation, invasion, and migration of CRC HCT116 cell line in vitro.
E2F1 protects CRC cells against cellular senescence
Oxidative stress and DDR promote cellular senescence (38). Moreover, E2F1 deficiency causes DNA replication damage (39) and p53-dependent apoptosis in various tumors. Based on the abovementioned experimental results, we hypothesized that E2F1 is a critical repressive regulator of cellular senescence in CRC.
TP53, cyclin dependent kinase inhibitor 1A (CDKN1A/p21), and cyclin-dependent kinase inhibitor 2A (CDKN2A/p16) (40) were used as the markers of the progression of cellular senescence, and their activation resulted in the initiation of apoptosis, autophagy, and other processes. We assessed the expression levels of these cellular senescence markers in the abovementioned cell lines using Western blot (Figure 3A). The results showed that all of them increased following the overexpression of E2F1, and the expression level of p-TP53 also increased dramatically. Furthermore, the senescent cells characteristically express β-galactosidase, which has high enzymatic activity at pH 6.0. We used senescence-associated β-galactosidase staining to determine the amounts of β-galactosidase expression in each stable cell line. The results revealed that β-galactosidase production was lower in the E2F1 overexpression group than in the Mock group; however, the E2F1 knockdown resulted in higher levels of β-galactosidase generated in the cells (Figure 3B). Furthermore, the HP1-γ protein is widely used to indicate tissue and cellular senescence as a marker for forming senescence-associated heterochromatin foci. Cellular immunohistochemistry confirmed that E2F1 knockdown promoted the expression of HP1-γ (red) and the formation of focal clumps in the nucleus (Figure 3C).
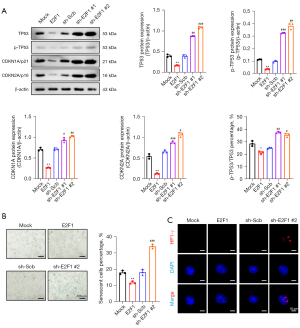
In summary, E2F1, a significant regulatory transcription factor of cellular senescence, suppressed the initiation of cellular senescence in CRC cells, whereas E2F1 deficiency resulted in premature senescence and consequent cell death.
E2F1 improves CRC resistance to oxaliplatin by suppressing cellular senescence
The function of cellular senescence in tumor chemotherapy has been shown previously (17,41); however, we wanted to know whether E2F1 affects chemotherapy by regulating cellular senescence in CRC. Thus, we created an oxaliplatin-resistant HCT116 cell line (OXA-HCT116) (42). MTT was used to assess the cell viability of OXA-HCT116 and HCT116 after oxaliplatin co-culture for 24 hours. The results showed that the survivability of OXA-HCT116 was substantially higher than that of HCT116, indicating that OXA-HCT116 was resistant to oxaliplatin (Figure 4A).
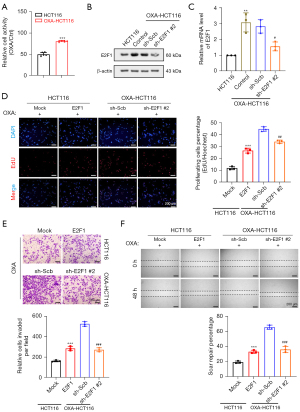
Western blot and qRT-PCR revealed that the E2F1 expression was higher in OXA-HCT116 than in HCT116, and sh-E2F1 #2 effectively reduced the E2F1 expression in OXA-HCT116 (Figure 4B,4C). Subsequently, we examined the importance of E2F1 in CRC chemoresistance. The proliferation, invasion, and migration of HCT116 cells were promoted by stable E2F1 overexpression and those of OXA-HCT116 cells were lessened by E2F1 knockdown (Figure 4D-4F).
After 24 hours of co-culture with oxaliplatin, the expression of senescence markers TP53 and p-TP53, CDKN1A/p21, and CDKN2A/p16 were significantly elevated in the HCT116 cells, and heterochromatic foci were observed in the nucleus (Figure 5A). Nevertheless, these alterations were removed after steady E2F1 overexpression. Moreover, the E2F1 knockdown of the OXA-HCT116 cells promoted senescence factors expression and heterochromatic foci formation; however, the control group cells did not show these alterations (Figure 5B,5C).
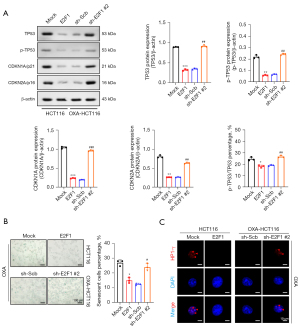
These findings suggested that oxaliplatin induces senescence in CRC cells, which was consistent with the previously reported results (43), and that E2F1 improves resistance to oxaliplatin by suppressing cell senescence.
Discussion
CRC is the third most prevalent cancer worldwide, whereas rectal cancer is typically only second behind colon cancer in fatality (44,45). The treatment of patients with cancer is difficult. First-line chemotherapeutic agents for CRC, such as capecitabine and bevacizumab, are frequently used in chemotherapy for CRC and are often combined with oxaliplatin in metastatic CRC chemotherapy (46). However, the treatment of CRC is hampered by primary drug resistance and patients with acquired drug resistance (47). The present study examined the probable mechanisms of medication resistance in CRC and provided the possibility of overcoming oxaliplatin resistance in CRC.
Cellular senescence is a permanent cell cycle that is stopped by a specific amount of stress. Acute senescence prevents cancer; inducing tumor cell senescence by drugs is also a treatment method (48,49). In the present study, we discovered that the CRC HCT116 cells treated with oxaliplatin-induced cell senescence (50) and inhibited proliferation, invasion, and migration. Furthermore, the OXA-HCT116 cells co-cultured with oxaliplatin showed considerably lower TP53, p21, and p16 than HCT116 cells treated similarly. These results made us consider whether drug resistance in CRC cells was attributable to the emergence of inhibitory senescence factors (51). The relationship between suppressed cellular senescence and tumor drug resistance has been reported in other tumors. For example, FOXM1 amplified the NBS1 expression and ATM phosphorylation, making breast cancer cells resistant to epirubicin-induced cellular senescence (52). When the Ras/Raf/MEK/ERK pathway was upregulated, the coregulation of the Ras/PI3K/PTEN/Akt/mTOR pathway enhanced drug resistance by inhibiting chemotherapy-induced cell senescence in the MCF-7 cell line (53).
E2F1, as a cell cycle regulator, plays a complicated role in regulating cell proliferation. E2F1 overexpression encourages senescence in normal cells, whereas E2F1 inhibition induces senescence in tumor cells (54-56). For instance, silencing PAX8 caused decreased E2F1 expression in tumor cells, which caused the G1/S cell cycle arrest and cellular senescence (57). c-MYC promoted enhanced E2F1 expression by activating USP2-AS1, thus delaying the cellular senescence process (58). In the present study, we observed that E2F1 knockdown induced cell senescence, along with increased expression levels of TP53, p21, and p16. Increased β-galactosidase staining positive cells and the formation of heterochromatin foci in the nucleus after E2F1 knockdown further supported this. In contrast, the overexpression of E2F1 reduced oxaliplatin-induced cellular senescence. Furthermore, oxaliplatin-resistant cells showed high levels of E2F1 expression but E2F1 knockdown induced a senescence-associated phenotype in oxaliplatin-treated OXA-HCT116 cells. We were amazed to discover that the resistance of colorectal tumor cells to oxaliplatin was closely associated with their expression levels of E2F1. E2F1 acts as a cellular senescence suppressor, preventing the development of senescence in drug-resistant cells, thus increasing the resistance of CRC cells to oxaliplatin.
The treatment of patients with cancer can be difficult, and the development of tumor resistance increases its difficulty. Combined chemotherapy or the combination of radiotherapy and chemotherapy significantly increases the burden on these patients (59); therefore, research on tumor drug resistance is important. The present study demonstrated the significance of E2F1 in the theory of cellular senescence regulating tumor drug resistance, indicating that E2F1 was no longer only a factor regulated by the classical senescence signaling pathway. The development of targeted drugs for E2F1 will successfully reduce the treatment dilemma of clinical drug-resistant patients.
Conclusions
Our study results on the mechanism of drug resistance formation in CRC could facilitate the development of corresponding chemotherapy regimens and drugs targeting related proteins. Furthermore, this was a preliminary study, so future studies will focus on whether other molecules synergize with E2F1 to promote drug resistance or whether cellular senescence is dominant in developing drug resistance in CRC.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81974280) and the Foundation of Health Commission of Hubei Province (No. WJ2021M251).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4054/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4054/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4054/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4054/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Wong MC, Ding H, Wang J, et al. Prevalence and risk factors of colorectal cancer in Asia. Intest Res 2019;17:317-29. [Crossref] [PubMed]
- Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol 2019;14:89-103. [Crossref] [PubMed]
- Garrett WS. The gut microbiota and colon cancer. Science 2019;364:1133-5. [Crossref] [PubMed]
- Kelly D, Yang L, Pei Z. Gut Microbiota, Fusobacteria, and Colorectal Cancer. Diseases 2018;6:109. [Crossref] [PubMed]
- Cai Z, Liu Q. Understanding the Global Cancer Statistics 2018: implications for cancer control. Sci China Life Sci 2021;64:1017-20. [Crossref] [PubMed]
- Gorgoulis V, Adams PD, Alimonti A, et al. Cellular Senescence: Defining a Path Forward. Cell 2019;179:813-27. [Crossref] [PubMed]
- Calcinotto A, Kohli J, Zagato E, et al. Cellular Senescence: Aging, Cancer, and Injury. Physiol Rev 2019;99:1047-78. [Crossref] [PubMed]
- Zhong G, Qin S, Townsend D, et al. Oxidative stress induces senescence in breast cancer stem cells. Biochem Biophys Res Commun 2019;514:1204-9. [Crossref] [PubMed]
- Venkatachalam G, Surana U, Clément MV. Replication stress-induced endogenous DNA damage drives cellular senescence induced by a sub-lethal oxidative stress. Nucleic Acids Res 2017;45:10564-82. [Crossref] [PubMed]
- Neault M, Couteau F, Bonneau É, et al. Molecular Regulation of Cellular Senescence by MicroRNAs: Implications in Cancer and Age-Related Diseases. Int Rev Cell Mol Biol 2017;334:27-98. [Crossref] [PubMed]
- Chatterjee S. Telomeres in health and disease. J Oral Maxillofac Pathol 2017;21:87-91. [Crossref] [PubMed]
- Wang G, Fu Y, Hu F, et al. Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway. Cell Death Dis 2017;8:e2607. [Crossref] [PubMed]
- Ou HL, Schumacher B. DNA damage responses and p53 in the aging process. Blood 2018;131:488-95. [Crossref] [PubMed]
- Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest 2018;128:1238-46. [Crossref] [PubMed]
- Wang L, Lankhorst L, Bernards R. Exploiting senescence for the treatment of cancer. Nat Rev Cancer 2022;22:340-55. [Crossref] [PubMed]
- St-Jean S, De Castro AC, Lecours M, et al. NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence. Cancers (Basel) 2021;13:4414. [Crossref] [PubMed]
- Wang N, Zhu C, Xu Y, et al. Negative Regulation of PTEN by MicroRNA-221 and Its Association with Drug Resistance and Cellular Senescence in Lung Cancer Cells. Biomed Res Int 2018;2018:7908950. [Crossref] [PubMed]
- El-Far AH, Darwish NHE, Mousa SA. Senescent Colon and Breast Cancer Cells Induced by Doxorubicin Exhibit Enhanced Sensitivity to Curcumin, Caffeine, and Thymoquinone. Integr Cancer Ther 2020;19:1534735419901160. [Crossref] [PubMed]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995;81:323-30. [Crossref] [PubMed]
- Fouad S, Hauton D, D'Angiolella V. E2F1: Cause and Consequence of DNA Replication Stress. Front Mol Biosci 2020;7:599332. [Crossref] [PubMed]
- Li SY, Zhu Y, Li RN, et al. LncRNA Lnc-APUE is Repressed by HNF4α and Promotes G1/S Phase Transition and Tumor Growth by Regulating MiR-20b/E2F1 Axis. Adv Sci (Weinh) 2021;8:2003094. [Crossref] [PubMed]
- Banerjee D, Gorlick R, Liefshitz A, et al. Levels of E2F-1 expression are higher in lung metastasis of colon cancer as compared with hepatic metastasis and correlate with levels of thymidylate synthase. Cancer Res 2000;60:2365-7.
- Rocca MS, Di Nisio A, Marchiori A, et al. Copy number variations of E2F1: a new genetic risk factor for testicular cancer. Endocr Relat Cancer 2017;24:119-25. [Crossref] [PubMed]
- Docquier A, Harmand PO, Fritsch S, et al. The transcriptional coregulator RIP140 represses E2F1 activity and discriminates breast cancer subtypes. Clin Cancer Res 2010;16:2959-70. [Crossref] [PubMed]
- Iglesias-Ara A, Zenarruzabeitia O, Buelta L, et al. E2F1 and E2F2 prevent replicative stress and subsequent p53-dependent organ involution. Cell Death Differ 2015;22:1577-89. [Crossref] [PubMed]
- Engelmann D, Pützer BM. Translating DNA damage into cancer cell death-A roadmap for E2F1 apoptotic signalling and opportunities for new drug combinations to overcome chemoresistance. Drug Resist Updat 2010;13:119-31. [Crossref] [PubMed]
- Ren B, Cam H, Takahashi Y, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 2002;16:245-56. [Crossref] [PubMed]
- Irwin M, Marin MC, Phillips AC, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 2000;407:645-8. [Crossref] [PubMed]
- Stanelle J, Pützer BM. E2F1-induced apoptosis: turning killers into therapeutics. Trends Mol Med 2006;12:177-85. [Crossref] [PubMed]
- Moiseeva O, Bourdeau V, Vernier M, et al. Retinoblastoma-independent regulation of cell proliferation and senescence by the p53-p21 axis in lamin A /C-depleted cells. Aging Cell 2011;10:789-97. [Crossref] [PubMed]
- Madan E, Gogna R, Kuppusamy P, et al. TIGAR induces p53-mediated cell-cycle arrest by regulation of RB-E2F1 complex. Br J Cancer 2012;107:516-26. [Crossref] [PubMed]
- Shen W, Song Z, Zhong X, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022;1:e36.
- Lachmann A, Xu H, Krishnan J, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010;26:2438-44. [Crossref] [PubMed]
- Zou Z, Ohta T, Miura F, et al. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res 2022;50:W175-82. [Crossref] [PubMed]
- Oki S, Ohta T, Shioi G, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 2018;19:e46255. [Crossref] [PubMed]
- Robinson AR, Yousefzadeh MJ, Rozgaja TA, et al. Spontaneous DNA damage to the nuclear genome promotes senescence, redox imbalance and aging. Redox Biol 2018;17:259-73. [Crossref] [PubMed]
- Choi EH, Kim KP. E2F1 facilitates DNA break repair by localizing to break sites and enhancing the expression of homologous recombination factors. Exp Mol Med 2019;51:1-12. [Crossref] [PubMed]
- Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol 2018;28:436-53. [Crossref] [PubMed]
- Childs BG, Gluscevic M, Baker DJ, et al. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov 2017;16:718-35. [Crossref] [PubMed]
- Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer 2019;18:43. [Crossref] [PubMed]
- Tomicic MT, Krämer F, Nguyen A, et al. Oxaliplatin-Induced Senescence in Colorectal Cancer Cells Depends on p14(ARF)-Mediated Sustained p53 Activation. Cancers (Basel) 2021;13:2019. [Crossref] [PubMed]
- Ciardiello D, Vitiello PP, Cardone C, et al. Immunotherapy of colorectal cancer: Challenges for therapeutic efficacy. Cancer Treat Rev 2019;76:22-32. [Crossref] [PubMed]
- Araghi M, Soerjomataram I, Jenkins M, et al. Global trends in colorectal cancer mortality: projections to the year 2035. Int J Cancer 2019;144:2992-3000. [Crossref] [PubMed]
- Benson AB, Venook AP, Al-Hawary MM, et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:329-59. [Crossref] [PubMed]
- Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017;8:59950-64. [Crossref] [PubMed]
- Wang Z, Gao J, Zhou J, et al. Olaparib induced senescence under P16 or P53 dependent manner in ovarian cancer. J Gynecol Oncol 2019;30:e26. [Crossref] [PubMed]
- Yang D, Zhang Q, Ma Y, et al. Augmenting the therapeutic efficacy of adenosine against pancreatic cancer by switching the Akt/p21-dependent senescence to apoptosis. EBioMedicine 2019;47:114-27. [Crossref] [PubMed]
- Qu K, Xu X, Liu C, et al. Negative regulation of transcription factor FoxM1 by p53 enhances oxaliplatin-induced senescence in hepatocellular carcinoma. Cancer Lett 2013;331:105-14. [Crossref] [PubMed]
- Zhou Y, Hu Y, Yang M, et al. The miR-106b~25 cluster promotes bypass of doxorubicin-induced senescence and increase in motility and invasion by targeting the E-cadherin transcriptional activator EP300. Cell Death Differ 2014;21:462-74. [Crossref] [PubMed]
- Khongkow P, Karunarathna U, Khongkow M, et al. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene 2014;33:4144-55. [Crossref] [PubMed]
- Taylor JR, Lehmann BD, Chappell WH, et al. Cooperative effects of Akt-1 and Raf-1 on the induction of cellular senescence in doxorubicin or tamoxifen treated breast cancer cells. Oncotarget 2011;2:610-26. [Crossref] [PubMed]
- Dimri GP, Itahana K, Acosta M, et al. Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 2000;20:273-85. [Crossref] [PubMed]
- Carey JP, Knowell AE, Chinaranagari S, et al. Id4 promotes senescence and sensitivity to doxorubicin-induced apoptosis in DU145 prostate cancer cells. Anticancer Res 2013;33:4271-8.
- Laine A, Sihto H, Come C, et al. Senescence sensitivity of breast cancer cells is defined by positive feedback loop between CIP2A and E2F1. Cancer Discov 2013;3:182-97. [Crossref] [PubMed]
- Li CG, Nyman JE, Braithwaite AW, et al. PAX8 promotes tumor cell growth by transcriptionally regulating E2F1 and stabilizing RB protein. Oncogene 2011;30:4824-34. [Crossref] [PubMed]
- Li B, Zhang G, Wang Z, et al. c-Myc-activated USP2-AS1 suppresses senescence and promotes tumor progression via stabilization of E2F1 mRNA. Cell Death Dis 2021;12:1006. [Crossref] [PubMed]
- Chang WH, Katsoulis M, Tan YY, et al. Late effects of cancer in children, teenagers and young adults: Population-based study on the burden of 183 conditions, in-patient and critical care admissions and years of life lost. Lancet Reg Health Eur 2022;12:100248. [Crossref] [PubMed]


