
Methylation factors as biomarkers of fibromyalgia
Highlight box
Key findings
• Fibromyalgia (FM) may be related to deoxyribonucleic acid methylation, and methylation factors, such as amino beta (A4) precursor protein binding family B member 2 (APBB2), A-kinase anchor protein 12 (AKAP12), and cluster of differentiation 38 (CD38), may be its potential biomarkers.
What is known and what is new?
• AKAP12 can be used as a diagnostic marker of FM, and CD38 and GATA binding protein 2 (GATA2) are also closely related to FM.
• Methylation factors amyloid beta precursor protein binding family b member 2 (APBB2), islet cell autoantigen 1 like (ICA1L), integrin subunit beta 8 (ITGB8) and kinesin light chain 3 (KLC3) may be potential biomarkers of FM.
What is the implication, and what should change now?
• This suggests that further exploration of methylation factors, such as APBB2, ICA1L, ITGB8 and KLC3, is needed to determine whether they can be used as biomarkers of FM.
Introduction
Fibromyalgia (FM) exists in 2–8% of the population, and is characterized by extensive pain, which is often accompanied by fatigue, memory problems, and sleep disorders (1,2). Although FM has been excluded from the diagnosis of neuropathic pain since the revision of the International Association for the Study of Pain (IASP) definition of neuropathic pain in 2011, recent studies have found that about 50% of FM patients have neuropathic changes of small and/or large fibers (3,4). One review reported an average prevalence of 2.7% (0.4–9.3%) worldwide, 3.1% in the Americas, 2.5% in Europe, and 1.7% in Asia (5). FM is a complex multisymptomatic long-term disease that places a heavy burden on healthcare systems around the world (6). FM is associated with billions of dollars of annual healthcare spending and a work disability rate of nearly 56% (7). FM lacks relevant specific laboratory tests and other ancillary tests, all tests and analyses will return normal results, and it has no known biomarkers (8). The etiology of FM remains unknown, and its diagnosis is clinical and not based on objective testing (9). Thus, little is known about how it can be effectively diagnosed and how clinicians and patients can use and understand biomarkers.
There are several factors related to the pathophysiology of FM, including central sensitization, the response of the hypothalamus pituitary adrenal axis to pressure, the promotion of inflammation, the reduction of anti-inflammatory cytokines, and interference with neurotransmitters, but the specific mechanism leading to FM is not yet clear (10).
Deoxyribonucleic acid (DNA) methylation mainly occurs in cytosine-C5 in the context of the cytosine-phosphate-guanosine (CpG) dinucleotide, which plays an important role in transcriptional regulation and genome stability (11). DNA methylation has been found in patients with FM (12). Related studies have shown that gene-environment interactions may be the trigger mechanism for FM: in particular, FM appears to be characterised by a pattern of DNA hypomethylation involving genes for stress responses, DNA repair, autonomic nervous system responses and subcortical neuronal abnormalities (13). In previous studies, DNA methylation has been found to be a biomarker for many diseases (14-16), while in FM, DNA methylation changes may reflect the role of immune-inflammatory responses and central sensitization (17). Therefore, the relationship between DNA methylation and FM needs to be further studied, which may help to evaluate and/or diagnose FM.
This study sought to explore the differential methylation factors between FM patients and control subjects, and identify the biological processes (BPs) involved in the differential methylation factors. By determining the epigenetic factors that form the pathophysiological basis of FM, we identified new indexes for the objective diagnosis of FM. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6631/rc).
Methods
Data sources
The data used in this study were downloaded from the Gene Expression Omnibus (GEO) database. The gene expression data of FM patients were obtained from the GSE67311 data set, which included 67 disease subjects and 75 control subjects. The methylation data were obtained from the GSE85506 data set, which included 24 disease subjects and 23 control subjects. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differential expression and methylation analyses
The raw data from the microarray data sets were normalized and log2-transformed. The differentially expressed genes (DEGs) from the GSE67311 data set were calculated with limma package (18). A P value <0.05 was set at the threshold. The differential methylation sites of the GSE85506 data set were calculated using the cAMP package (19). Subsequently, genes differentially expressed in contrast to methylation are used as methylation factors.
Functional enrichment analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the methylation factors were conducted using clusterProfiler package (20). A gene set enrichment analysis (GSEA) (21) of the methylation factors was carried out with GSEA software. A P value <0.05 was considered statistically significant.
Prediction of TFs
First, all the transcription factors (TFs) that regulate the methylation factors were obtained from TRRUST version 2 (22) human TF target data. The TFs with P values <0.05 according to the hypergeometric tests were used as the transcriptional regulators. All corresponding regulatory relationships in National Center for Biotechnology Information (NCBI) were then screened.
Receiver operating characteristic (ROC) curve analysis
The area under the curve (AUC) values of all the methylation factors were calculated and ROC curves were drawn. Genes with AUC values >0.7 were considered key methylation factors. In addition, the rms package was used to further construct the nomogram and calibration curve to evaluate the ability of key methylation factors to predict FM risk.
Assessment of cell deaths
A GSEA was performed using the clusterProfiler package to assess FM-related cell death (pyroptosis, necroptosis, and cuproptosis). Cell death was quantified by a gene set variation analysis (23). Pearson’s correlation coefficients were then used to assess the correlation between genes and cell death, and P value <0.05 was considered statistically significant.
Statistical analysis
For all bioinformatics analysis, P value <0.05 was considered statistically significant.
Results
FM-related methylation factors
To identify the disordered molecules related to FM, we analyzed the DEGs in the GSE67311 data set. A total of 1,787 DEGs were found, which included both upregulated and downregulated genes (Figure 1A). To further identify the methylation factors, the different methylation sites of GSE85506 were analyzed. A total of 9,015 methylation genes were identified from the 19,982 methylation sites (Figure 1B). After examining the relationship between methylation modification and gene expression, 455 methylation factors were identified (Figure 1C). These methylation factors may play an important role in FM.

Biological effects of methylation factors
Changes in the methylation spectrum between FM patients and normal control subjects may reveal important biological functions. The enrichment analysis showed that methylation factors were significantly involved in 599 BPs, 76 cell components (CCs), 151 molecular functions (MFs), and 9 KEGG signaling pathways. The statistical analysis showed that methylation factors are mainly involved in the regulation of neuron project development, the regulation of small molecular metallic process, and other biological functions (Figure 2A-2C). Moreover, methylation factors are mainly involved in the immune inflammation and metabolism related KEGG pathway (Figure 2D). In addition, the GSEA identified four KEGG signalling pathways involving the methylation factors, including the neural system and pain response–related pathways (Figure 2E).
TFs regulate methylation factors
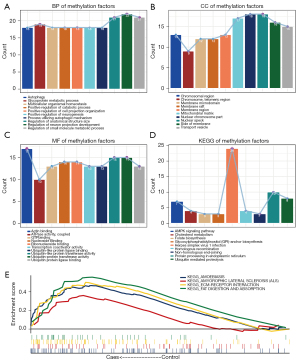
To identify the methylated integrated regulatory network of FM, we predicted the regulatory TFs of the methylated factors. After filtering the regulatory relationships reported in NCBI, a total of 18 TFs were found to regulate the methylation factors (Figure 3A). Among these, specificity protein 1 (SP1) and signal transducer and activator of transcription 3 (STAT3) regulate most of the methylation factors. In addition, we found a strong coupling relationship between TFs, particularly between SP1 and metastasis associated 1 (MTA1), suggesting that these two genes may play a synergistic regulatory role in the induction of FM (Figure 3B).
Identify key methylation factors
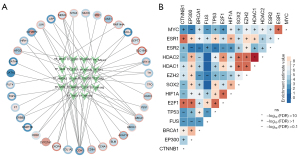
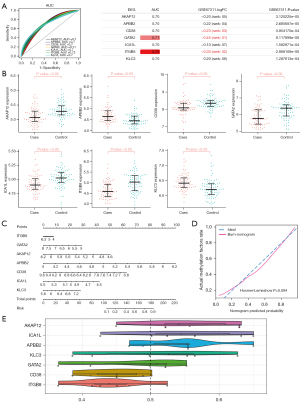
To identify the methylation factors with a clinical significance, the AUC values were calculated. The AUC values of 7 methylated genes [i.e., A-kinase anchor protein 12 (AKAP12), amino beta (A4) precursor protein binding family B member 2 (APBB2), cluster of differentiation 38 (CD38), GATA binding protein 2 (GATA2), islet cell autoantigen 1 like (ICA1L), integrin subunit beta 8 (ITGB8), and kinesin light chain 3 (KLC3)] were >0.7, and thus have high clinical diagnostic ability (Figure 4A). We also observed the expression of these genes in the GSE67311 data set (Figure 4B). The disease risk prediction ability of the 7 methylation factors was further evaluated by a nomogram (Figure 4C). Among them, APBB2, AKAP12 and CD38 showed better predictive power compared to other methylation factors and could be further investigated as potential biomarkers for FM. The calibration chart showed that the nomogram performed well compared to the ideal model (Figure 4D). The GO functions of APBB2 and AKAP12 are very similar, suggesting that they may be involved in similar BPs in affecting FM (Figure 4E).
Cell deaths influenced by key methylation factors
To evaluate whether cell death is present in FM patients, we performed a GSEA. The results showed that pyroptosis, necroptosis, and cuproptosis were significantly inhibited in FM (Figure 5A). The correlation results revealed a significant negative correlation between AKAP12 and all 3 types of cell death, and a significant negative correlation between CD38 and cell death (Figure 5B).

Discussion
In this study, we used bioinformatics methods and identified a set of potential molecular diagnostic markers for FM. Currently, the diagnosis of FM is based solely on a comprehensive clinical assessment and no validated biomarkers associated with FM have been identified. Therefore, the FM biomarkers reported here are of great clinical significance. In particular, our study showed that the 7 differential methylation factors in the peripheral blood of the FM patients had good clinical diagnostic ability. Notably, transcriptomic studies have shown that between 35% and 80% of known transcripts are present in brain and blood tissue samples (24), and therefore blood samples can be a reliable and easy source of FM biomarkers. Some studies had measured the excitability parameters of the cerebral cortex in FM patients and control subjects and found that the results were parallel to the changes in the peripheral blood methylation levels in FM patients (12). These findings highlight the importance of peripheral blood DNA methylation in future FM biomarker studies.
In the whole life process, the expression of genes changes are partly affected by epigenetic mechanisms, such as DNA methylation (25). Among the seven key methylation factors we identified, the APBB2 gene is known to be related to Alzheimer’s disease, which affects the cognitive ability of patients (26,27). In this study, APBB 2 had low methylation and high expression, which may be related to the memory and sleep problems of FM patients. AKAP12 is a new and effective scaffold protein, which plays an important role in protein kinase C, protein kinase A, cyclin, F-actin, and other key signal factors (28). AKAP12 gene knockout has been shown to promote the infiltration of inflammatory cells, the production of reactive oxygen species, and autophagy (29). The low expression of AKAP12 in FM patients may lead to the increase of pro-inflammatory cytokines, which was related to the pathophysiology of FM (30). Moreover, CD 38 is also considered a candidate gene for FM (31). The absence of CD 38 will lead to the development of astrocytes and oligodendrocytes in mice and affect the function of the central nervous system (32).
In addition, by predicting transcription regulators, we obtained the key regulator SP1 and STAT3. SP1 can regulate gene expression positively and negatively by binding to CpG rich sites, and is affected by the methylation status of these regions (33). STAT3 plays a key role in inflammation and immune control and mutations in it have been associated with diseases such as immunodeficiency, autoimmunity and cancer (34). It is suggested that SP1 and STAT3 may participate in the KEGG pathway related to immune inflammation and metabolism by regulating methylation factors, thus affecting the development of FM.
The destruction of the epigenetic mechanism is related to various immune, neurological, and endocrine diseases (35). In this study, the enrichment analysis of the methylated factors revealed more biological functions and signaling pathways related to the nerves, immune inflammation, and metabolism. Previous studies had shown that the central nervous system may play an important role in the occurrence and maintenance of FM (36). In FM patients, pain is not only related to noxious stimulation, but is also related to central pain management abnormality (37). The pathophysiology of FM is not clear, but it may involve the immunoinflammatory pathway (38). Metabonomic results have shown that the levels of arginine and ornithine in patients with FM are increased (39). Interlukin-1β triggers NLR family pyrin domain containing 3 (NLRP3) inflammasome activation, inducing inflammatory cell death, which in turn participates in chronic pain responses (40). These results suggested that the differential methylation of genes play an important role in the development and maintenance of the nervous system, immune inflammation and pain response, and may be an important factor in FM.
This study provided a new model for understanding the dynamic interactions among the stochastic, environmental, genetic, and epigenetic factors that influence pain perception and expression in FM patients by examining the patterns of DNA methylation modifications that regulate gene expression profiles. Our results may serve as a starting point for further large-scale and independent population-based research on DNA methylation as a possible disease mechanism for FM. Moreover, an understanding of the key methylation factors of FM may lead to new diagnosis and treatment options. However, the sample included in our study analysis was relatively small, so the findings need to be analyzed in a larger sample and further validated using experiments.
Conclusions
Our study suggests that FM is associated with DNA methylation. The methylation factors APBB2, AKAP12, and CD38 should be further investigated as potential biomarkers. Our findings may provide a new biological framework for the possible disease mechanisms underlying FM.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6631/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6631/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The expression profiles used in this study was downloaded from the GEO database (access Nos. GSE67311, GSE85506). The corresponding ethical approval and informed consent of patients have been provided in the database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Assavarittirong C, Samborski W, Grygiel-Gorniak B. Oxidative Stress in Fibromyalgia: From Pathology to Treatment. Oxid Med Cell Longev 2022;2022:1582432. [Crossref] [PubMed]
- Alvarez MC, Albuquerque MLL, Neiva HP, et al. Understanding the Associations across Fibromyalgia-Related Fatigue, Depression, Anxiety, Self-Esteem Satisfaction with Life and Physical Activity in Portuguese and Brazilian Patients: A Structural Equation Modeling Analysis. Medicina (Kaunas) 2022;58. [Crossref] [PubMed]
- Cheng CW, Wong CS, Hui GK, et al. Fibromyalgia: is it a neuropathic pain? Pain Manag 2018;8:377-88. [Crossref] [PubMed]
- Viceconti A, Geri T, De Luca S, et al. Neuropathic pain and symptoms of potential small-fiber neuropathy in fibromyalgic patients: A national on-line survey. Joint Bone Spine 2021;88:105153. [Crossref] [PubMed]
- Welsch P, Uceyler N, Klose P, et al. Serotonin and noradrenaline reuptake inhibitors (SNRIs) for fibromyalgia. Cochrane Database Syst Rev 2018;2:CD010292. [Crossref] [PubMed]
- Wilson N, Beasley MJ, Pope C, et al. UK healthcare services for people with fibromyalgia: results from two web-based national surveys (the PACFiND study). BMC Health Serv Res 2022;22:989. [Crossref] [PubMed]
- Walitt B, Katz RS, Bergman MJ, et al. Three-Quarters of Persons in the US Population Reporting a Clinical Diagnosis of Fibromyalgia Do Not Satisfy Fibromyalgia Criteria: The 2012 National Health Interview Survey. PLoS One 2016;11:e0157235. [Crossref] [PubMed]
- Perrot S. Fibromyalgia: A misconnection in a multiconnected world? Eur J Pain 2019;23:866-73. [Crossref] [PubMed]
- Pastor-Mira MA, Lopez-Roig S, Toribio E, et al. Pain-Related Worrying and Goal Preferences Determine Walking Persistence in Women with Fibromyalgia. Int J Environ Res Public Health 2022;19. [Crossref] [PubMed]
- Uceyler N, Burgmer M, Friedel E, et al. Etiology and pathophysiology of fibromyalgia syndrome: Updated guidelines 2017, overview of systematic review articles and overview of studies on small fiber neuropathy in FMS subgroups. Schmerz 2017;31:239-45. [Crossref] [PubMed]
- Wright DFB, Martin JH, Cremers S. Spotlight Commentary: Model-informed precision dosing must demonstrate improved patient outcomes. Br J Clin Pharmacol 2019;85:2238-40. [Crossref] [PubMed]
- Ciampi de Andrade D, Maschietto M, Galhardoni R, et al. Epigenetics insights into chronic pain: DNA hypomethylation in fibromyalgia-a controlled pilot-study. Pain 2017;158:1473-80. [Crossref] [PubMed]
- D'Agnelli S, Arendt-Nielsen L, Gerra MC, et al. Fibromyalgia: Genetics and epigenetics insights may provide the basis for the development of diagnostic biomarkers. Mol Pain 2019;15:1744806918819944. [Crossref] [PubMed]
- Muller D, Gyorffy B. DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim Biophys Acta Rev Cancer 2022;1877:188722. [Crossref] [PubMed]
- Shirvani-Farsani Z, Maloum Z, Bagheri-Hosseinabadi Z, et al. DNA methylation signature as a biomarker of major neuropsychiatric disorders. J Psychiatr Res 2021;141:34-49. [Crossref] [PubMed]
- Luo C, Huang J, Guo Z, et al. Methylated biomarkers for breast cancer identified through public database analysis and plasma target capture sequencing. Ann Transl Med 2021;9:683. [Crossref] [PubMed]
- Gerra MC, Carnevali D, Ossola P, et al. DNA Methylation Changes in Fibromyalgia Suggest the Role of the Immune-Inflammatory Response and Central Sensitization. J Clin Med 2021;10:4992. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Arumugham VB, Baldari CT. cAMP: a multifaceted modulator of immune synapse assembly and T cell activation. J Leukoc Biol 2017;101:1301-16. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Han H, Cho JW, Lee S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res 2018;46:D380-D6. [Crossref] [PubMed]
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain "-omes Am J Med Genet B Neuropsychiatr Genet 2013;162B:595-603. [Crossref] [PubMed]
- Livshits G, Malkin I, Freidin MB, et al. Genome-wide methylation analysis of a large population sample shows neurological pathways involvement in chronic widespread musculoskeletal pain. Pain 2017;158:1053-62. [Crossref] [PubMed]
- Readhead B, Haure-Mirande JV, Funk CC, et al. Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 2018;99:64-82 e7. [Crossref] [PubMed]
- Golanska E, Sieruta M, Gresner SM, et al. APBB2 genetic polymorphisms are associated with severe cognitive impairment in centenarians. Exp Gerontol 2013;48:391-4. [Crossref] [PubMed]
- Soh RYZ, Lim JP, Samy RP, et al. A-kinase anchor protein 12 (AKAP12) inhibits cell migration in breast cancer. Exp Mol Pathol 2018;105:364-70. [Crossref] [PubMed]
- Li Y, Yu QH, Chu Y, et al. Blockage of AKAP12 accelerates angiotensin II (Ang II)-induced cardiac injury in mice by regulating the transforming growth factor beta1 (TGF-beta1) pathway. Biochem Biophys Res Commun 2018;499:128-35. [Crossref] [PubMed]
- Derry S, Wiffen PJ, Hauser W, et al. Oral nonsteroidal anti-inflammatory drugs for fibromyalgia in adults. Cochrane Database Syst Rev 2017;3:CD012332. [Crossref] [PubMed]
- Qiu Y, Zhang TJ, Meng LB, et al. Bioinformatics analysis of gene and microRNA targets for fibromyalgia. Clin Exp Rheumatol 2021;39:21-31. [Crossref] [PubMed]
- Hattori T, Kaji M, Ishii H, et al. CD38 positively regulates postnatal development of astrocytes cell-autonomously and oligodendrocytes non-cell-autonomously. Glia 2017;65:974-89. [Crossref] [PubMed]
- Li L, He S, Sun JM, et al. Gene regulation by Sp1 and Sp3. Biochem Cell Biol 2004;82:460-71. [Crossref] [PubMed]
- Hillmer EJ, Zhang H, Li HS, et al. STAT3 signaling in immunity. Cytokine Growth Factor Rev 2016;31:1-15. [Crossref] [PubMed]
- Trivedi MS, Oltra E, Sarria L, et al. Identification of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome-associated DNA methylation patterns. PLoS One 2018;13:e0201066. [Crossref] [PubMed]
- Kong J, Wolcott E, Wang Z, et al. Altered resting state functional connectivity of the cognitive control network in fibromyalgia and the modulation effect of mind-body intervention. Brain Imaging Behav 2019;13:482-92. [Crossref] [PubMed]
- Vanneste S, Ost J, Van Havenbergh T, et al. Resting state electrical brain activity and connectivity in fibromyalgia. PLoS One 2017;12:e0178516. [Crossref] [PubMed]
- Andrés-Rodríguez L, Borràs X, Feliu-Soler A, et al. Machine Learning to Understand the Immune-Inflammatory Pathways in Fibromyalgia. Int J Mol Sci 2019;20:4231. [Crossref] [PubMed]
- Clos-Garcia M, Andres-Marin N, Fernandez-Eulate G, et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine 2019;46:499-511. [Crossref] [PubMed]
- Starobova H, Nadar EI, Vetter I. The NLRP3 Inflammasome: Role and Therapeutic Potential in Pain Treatment. Front Physiol 2020;11:1016. [Crossref] [PubMed]


