
Use of exosome transcriptome-based analysis to identify novel biomarkers in patients with locally advanced esophageal squamous cell carcinoma undergoing neoadjuvant chemoradiotherapy
Highlight box
Key findings
• Two miRNA-mRNA networks (IFIT2-miR-3615-IFIT2-miR-484 and BTN3A3-miR-6803-3p) were identified as predictive biomarkers in patients with esophageal squamous cell carcinoma who have received neoadjuvant chemoradiotherapy.
What is known and what is new?
• A profile of an exosome transcriptome in patients with esophageal cancer who have received surgery alone has been established in the existing bioinformatics database.
• This present exosome transcriptome-based analysis revealed novel biomarkers to predict treatment response in patients with locally advanced esophageal squamous cell carcinoma undergoing neoadjuvant chemoradiotherapy.
What is the implication, and what should change now?
• Our results expanded upon the expression profile of exosome transcriptome in patients with esophageal squamous cell carcinoma who have received neoadjuvant chemoradiotherapy, which could be applied to predict treatment response after further confirmation.
Introduction
Esophageal cancer is one of the most aggressive malignancies of the gastrointestinal tract, with a 5-year survival rate of less than 25% (1). In China, esophageal squamous cell carcinoma (ESCC) is the most commonly observed malignant type, with Chinese cases accounting for more than half of the global burden. Neoadjuvant chemoradiotherapy (nCRT) followed by surgery is currently still the major treatment strategy for locally advanced ESCC and was demonstrated to improve survival by the landmark CROSS and NEOCRTEC5010 studies (2,3). Despite the survival benefits, half of patients still relapse after nCRT and surgery, especially those without pathologic complete response (pCR). This suggests that more effective biomarkers are required to increase the accuracy of predicting pCR to identify the patients most suited to receive nCRT.
Liquid biopsy-based biomarkers in cancer diagnostics are associated with low cost and minimal invasiveness (4). Significant efforts have been made in the use of liquid biopsy for the early detection and treatment stratification of cancer, as well as residual disease and recurrence monitoring. Although circulating tumor cells and circulating tumor DNA have been used for early detection of tumors, exosomes and other extracellular vesicles have become a platform with potentially wider and complementary applications (5). The exosomes secreted by tumor cells consist of many molecules closely related to the characteristics of tumor. Analysis of exosomes is benefit to the diagnose of tumor and prediction of the tumor prognosis (6). Previous study has shown that the exosomes secreted by tumors can enter into circulatory system and be detected in the blood (7). Separating the exosomes in the blood could realize the non-invasive way to identify the characteristics of tumors at an early stage and potential therapeutic targets. Therefore, plasma exosomes are attractive as biomarkers for disease progression and risk stratification for applying nCRT in patients with ESCC.
Exosomes have been reported to promote tumorigenesis in several cancers, particularly through transfer of miRNAs. Circulating exosome-miRNAs (exo-miRNAs) have been suggested as specific and stable molecular biomarkers (8,9). Besides miRNAs, the combination of other RNA types [i.e., messenger RNA (mRNA), long noncoding RNA (lncRNA), and circulating RNA (cRNA)] can provide more comprehensive information and a reference for the molecular mechanism regarding nCRT. Exosome transcriptome could represent a more detailed and specific molecular biomarker compared with a single RNA type. The use of exosome transcriptome to predict the treatment response of nCRT has been widely used in other cancers (such as breast cancer and gastric cancer) (10,11); however, it is still in its infancy and has not been reported in ESCC. In addition, the current efficacy prediction mainly uses exo-miRNA panels, which cannot reveal the deep relationship between the three important types of exo-RNAs (mRNA, miRNA, and lncRNA). Therefore, we aimed to determine the associations between miRNA, mRNA, and lncRNA expression and nCRT response in patients with ESCC. To the best of our knowledge, this is the first study to use the exosome transcriptome with three types of RNA to examine the biomarkers in patients with ESCC who received nCRT and surgery. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-23-452/rc).
Methods
Patients and specimens
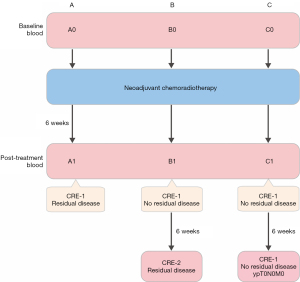
The patient population consisted of 15 patients with ESCC who received nCRT and surgery in Shanghai Chest Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) (12). This study was approved by the institution ethics committee of Shanghai Chest Hospital (No. KS2160). All patients provided written informed consent for the use of blood and specimens. The study flowchart is shown in Figure 1. The CROSS regimen (i.e., 5 weekly cycles of carboplatin and paclitaxel with concurrent radiation) was used in nCRT. Previous study showed that roughly 40% of patients with ESCC have a pathologically complete response (pCR) in the resection specimen after nCRT (3). For these patients surgical resection might not be necessary. Hence, an active surveillance strategy has been proposed in which patients will undergo frequent clinical response evaluations. We conducted a second response to increase the diagnostic accuracy and therefore patients will be divided into three groups. For each patient, a blood sample was collected (time point 0) before nCRT. The clinical response was evaluated at 6 weeks after nCRT, and another blood sample was collected (time point 1). Subsequently, radical esophagectomy was performed. According to the pathologic results, patients were divided into 3 groups. For group A, the clinical assessment observed residual tumor, and this was confirmed by the surgery. For group B, no residual disease was observed after nCRT, but tumor progression was discovered after 6-week follow-up. For group C, complete tumor remission was achieved (ypT0N0M0). Five patients in each group were selected (with similar baseline information), and for each patient, 2 blood samples at different time points of the same patient were sent for exosome purification and sequencing.
Exosomal RNA purification
The exosomes were isolated from the blood plasma samples. Quality control were indicated with representative images of transmission electron microscopy (Figure S1) and western blotting for positive markers of exosomes (Figure S2). The diluted serum specimens were centrifuged (500 g for 5 min and 2,000 g for 10 min). The supernatant was collected and centrifuged (10,000 g) for another 30 min. The pellet was filtered with a 0.22-µm filter and sent for ultracentrifugation (110,000 g for 90 min). The resuspended solution was later ultracentrifuged for another 90 min at 110,000 g. After removal of the supernatant, resuspension of the final pellet (with phosphate-buffered saline (PBS)] was later concentrated at 4,000 g using the MilliporeAmicon Ultra-15 Centrifugal Filter Unit. When the volume reached 100 µL, the process was stopped. The concentrated exosomes were placed at −80 ℃. During exosome analysis, the particle size of nanoparticle suspension was examined using scattered light detection. Then, the size and structure of exosomes were directly observed under transmission electron microscopy. Plasma exosomes were further confirmed with following protein markers under Western blotting: positive for CD9/TSG101/HSP70 and negative for calnexin. RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit (Thermo Fisher, Inc. Waltham, MA, USA). Amplification and complement DNA (cDNA) labeling were performed using the SensationPlus FFPE Amplification Kit (Thermo Fisher Scientific) according to manufacturer’s instructions.
Gene expression analysis
Raw reads in FastQ format were acquired using the Illumina HiSeq2500 platform. Clean data were obtained by removing low-quality reads and those containing adapter or ploy-N. For miRNA analysis, the filtered ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and other ncRNA and repeats were removed. The remaining reads were used to detect known miRNAs and novel miRNAs as predicted via the comparison with the known miRNAs from miRBase. For lncRNA analysis, the HISAT2 software was used. Sequence alignment and subsequent analysis were performed based on the GRCh38 reference. After alignment, the reads were assembled using StringTie software. The mRNA, miRNA, and lncRNA expression profile were analyzed for differentially expressed genes (DEGs). According to the grouping information, the sample expression data were imported into the “DEGseq” R package (The R Foundation for Statistical Computing) to compare the expression differences between samples. The fold change (FC) and adjusted P value were obtained for each transcript. Then, according to the threshold of P value <0.05 and |log2(FC)| ≥0.585, the DEGs of each data set were acquired. Volcano plots were produced for DEGs and DE transcripts using the “ggplot2” R package. In particular, we paid attention to the DE mRNAs/lncRNAs and miRNAs with opposite change directions. The intersection of different sets was visualized in a Venn diagram. For each comparison, a heatmap of DEGs between 2 groups was drawn.
Target prediction of key miRNAs and screening of key oncogenes and tumor-suppressor genes
The mRNAs potentially targeted by key miRNAs were predicted using the miRWalk online tool (http://mirwalk.umm.uni-heidelberg.de/). The following pairs were specifically scrutinized: tumor-promoting miRNAs, tumor-suppressor mRNAs, tumor-suppressor miRNAs, and tumor-promoting mRNAs.
Enrichment analysis
Based on key oncogenes and tumor-suppressor genes, we used the Metascape tool to explore the enriched Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Reactome sets, canonical pathways, TRRUST sets, and enriched transcription factor-targets. Terms with a P value <0.05, a minimum count of 3, and an enrichment factor >1.5 were collected.
Protein-protein interaction (PPI) analysis
Using all the key genes, PPI enrichment analysis was conducted based on Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/). All interactions except text mining (physical score >0.14) were used to establish the PPI network.
Results
Remission- and progression-associated DEGs (mRNAs)
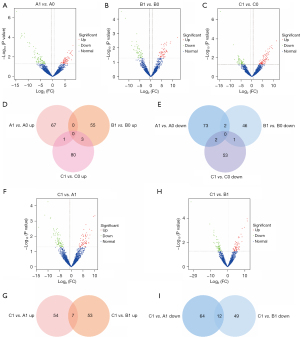
First, we compared the exosome transcriptome at 2 time points (before and after nCRT) in 3 cohorts (n=5 per group). In group A, there were 73 upregulated mRNAs and 184 downregulated mRNAs (Figure 2A); in group B, there were 68 upregulated and 26 downregulated ones (Figure 2B); in group C, there were 118 upregulated and 26 down-regulated ones (Figure 2C). The intersection of upregulated and downregulated genes was visualized in a Venn diagram (Figure 2D). Theoretically, the 74 unique upregulated genes in the C1 vs. C0 comparison (but not B1 vs. B0 or A1 vs. A0) were associated with nCRT response or tumor remission (Figure 2D, named only-C-Up set); the unique downregulated genes in the C1 vs. C0 comparison (but not B1 vs. B0 or A1 vs. A0) were associated with ESCC progression (Figure 2D, named only-C-Down set). Among the common genes between A1-A0 DEGs and B1-B0 DEGs, the unique upregulated gene (not among C1-C0 DEGs) appeared to be associated with ESCC progression (Figure 2D, named only-AB), while the unique downregulated gene (not among C1-C0 DEGs) appeared to be associated with ESCC remission (Figure 2D, named only-AB-Down set). The significance of these 4 sets is particularly important in nCRT treatment.
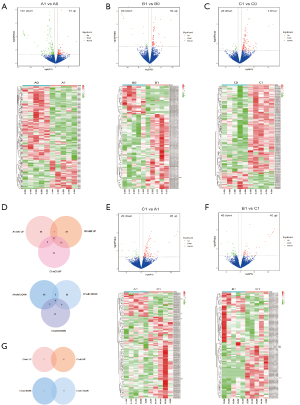
Next, at the second time point, the DEGs of C1 vs. A1 and C1 vs. B1 were acquired, including 95 C1-A1 upregulated (Figure 2E) and 45 C1-B1 upregulated (Figure 2F) DEGs. The intersection of C1-A1 upregulation and C1-B1 upregulation included 3 genes (Figure 2G, named CvsAB-Up) potentially associated with remission, and the intersection of C1-A1 downregulation and C1-B1 downregulation included 1 gene (Figure 2G, named CvsAB-Down) potentially associated with progression. Overall, ESCC remission was associated with 3 sets: only-C-Up, only-AB-Down, and CvAB-Up, while ESCC progression was associated with another 3 sets: only-C-Down, only-AB-Up, and CvAB-Down.
Enrichment analysis of ESCC progression and remission genes
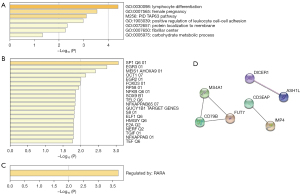
In the progression gene enrichment, we removed unknown genes and input the remaining 23 genes. In GO enrichment (Figure 3A), the following terms are enriched: lymphocyte differentiation, female pregnancy, and positive regulation of leukocyte cell-cell adhesion, among others. In enriched the canonical pathways, the TAP63 pathway was enriched. In the transcription-factor targets, the following factors appeared enriched: SP1, EGR3, MEIS1AHOXA9, OCT1, EGR2, FOXD3, RP58, NFKB, and SOX9, among others (Figure 3B). The TRRUST enrichment showed that RARA may modulate these progression genes (Figure 3C). Based on these 23 genes, a PPI network showed that MS4A1, CD79B, FUT7, CD3EAP, IMP4, DICER1, and ASH1L were located at the key positions, which indicated the crucial role of these genes in ESCC progression (Figure 3D).
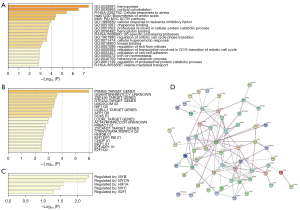
Similarly, 76 remission-associated genes were enriched in many GO terms, including in hemopoiesis, cortical cytoskeleton, and cellular response to leukemia inhibitory factor (Figure 4A). Reactome analysis also indicated that up-specific processing proteases, RUNX1 regulates transcription of genes involved in differentiation of hematopoietic stem cells (HSCs), vesicle-mediated transport, and others, were enriched. Meanwhile, biosynthesis of amino acids and carbon metabolism were the enriched KEGG pathways. In the transcription factor-target analysis, the following factors were enriched: PSMB5, ZNF436, GTF2A2, COBLL1, AFP1, and SOX5, among others (Figure 4B); and remission associated genes appeared to be largely regulated by MYB, MYCN, HIF1A, MYC, and E2F1 (Figure 4C). The PPI network of the remission-associated genes is shown as Figure 4D, in which UBB, IFIT1B, AHSP, MAT2A, SELENBP1, SPTA1, and TNS1 appear as the hub genes. Taken together, these results showed that the filtered hub genes play an important role in tumorigenesis and development.
Remission and progression associated miRNAs
The DE miRNAs were analyzed in the same protocol. The altered miRNAs in A1 vs. A0, B1 vs. B0, and C1 vs. C0 are shown in Figure 5A-5D. In Figure 5E-5G, there are 3 miRNAs in the only-C-Up set and 5 miRNAs in the only-C-Down set (hsa-miR-92b-3p, hsa-miR-6803-3p, hsa-miR-484, hsa-miR-941, and hsa-miR-3615 are within the only-C-Down set; hsa-miR-122-5p is within the CvsAB-Down set; hsa-miR-335-5p and hsa-miR-132-3p are within the only-C-Up set; and there are no known miRNAs in the only-AB-Up, only-AB-Down, or CvsAB-Up sets).
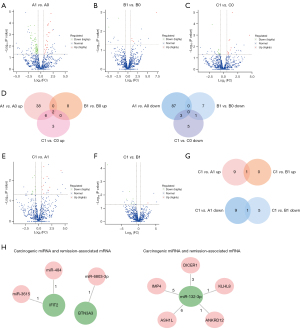
Key miRNA-mRNA pairs
The targets of remission- and progression-associated miRNAs were then screened. Between the remission-associated mRNA and the targets of progression-associated (carcinogenic) miRNAs, the intersection was acquired, and 2 miRNA-mRNA networks were identified (Figure 5H): IFIT2-miR-3615-IFIT2-miR-484 and BTN3A3-miR-6803-3p. Meanwhile, we screened the intersection of progression-associated (carcinogenic) mRNAs and the targets of remission-associated miRNAs. Based on network analysis, miR-132-3p (remission-associated miRNA) was located at the core and matched with DICER1, KLHL8, ANKRD12, ASH1L, and IMP4.
Remission- and progression-associated lncRNAs
Finally, the DE lncRNAs were analyzed. There were 80 lncRNAs in the only-C-Up set, 2 lncRNAs in the only-AB-Down set, and 53 lncRNAs in the only-C-Down set (Figure 6A-6E). Additionally, there were 61 C1-A1 upregulated lncRNAs and 60 C1-B1 upregulated lncRNAs, between which 7 DE lncRNAs were shared (Figure 6F,6G). Meanwhile, we discovered 76 C1-A1 downregulated lncRNAs and 61 C1-B1 downregulated lncRNAs, between which 12 lncRNAs were shared (Figure 6H,6I). For these remission- and progression-associated lncRNAs, we screened all the cis-target genes and trans-target genes, but no common ones were found with the remission- and progression-associated mRNAs. Finally, the lncRNA targets of key miRNAs (as mentioned above) were collected, but no common ones were shared with these remission- and progression-associated lncRNAs.
Discussion
The significant transcriptomes associated with ESCC have been widely investigated through high-throughput approaches. For example, known ESCC-related miRNAs include miR-23b-5p, miR-877-3p, and miR-17-92, among others (13-16). Several published works demonstrated that chemotherapy resistance can be affected by mRNAs like TUG1, ZFX, and NRF2, and miRNAs like miR-130a-3p, miR-125a-5p, and miR-224 (17-21). However, the key prognostic molecules in the therapeutic mechanism of ESCC, especially in nCRT, still need to be investigated. In this study, we used transcriptome analysis to assess the global signature of exo-RNAs and identified distinct exo-RNA signatures that may effectively predict treatment outcomes of nCRT. Here, CCDC85B (only-AB-Up) and stearoyl-CoA desaturase (SCD) (CvsAB-Down) were found to be 2 important progression-associated mRNAs, while IFIT2 was found to be a key remission-associated mRNA (CvsAB-Up). Additionally, several important miRNA-mRNA networks were identified: miR-484-IFIT2, miR-3615-IFIT2, miR-6803-3p-BTN3A3, and miR-132-3p-DICER1-KLHL8-ANKRD12-ASH1L-IMP4. All these findings are highly novel, as the roles of these molecules in the development of ESCC and treatment response to nCRT are completely unknown or not well studied.
In colorectal cancer, CCDC85B (also known as DIPA) is a carcinogenetic gene (22). It could be found on the centrosome and directly interacts with p78 and influences malignant transformation by modulating gene transcription (23). It has been reported that CCDC85B is a master regulator that can transform the cellular state of fast-growing subtype cells into the slow-growing subtype (24). SCD is a member of the de novo fatty-acid synthesis pathway (25), and it has been associated with esophageal cancer (26). Additionally, we found IFIT2 to be a key remission-associated mRNA, and the consistent result of the miR-484-IFIT2 axis was a particularly striking finding in this work. Currently, the direct association between ESCC and IFIT2 has not been clearly reported. IFIT2 (interferon-induced protein with tetratricopeptide repeats 2) enables RNA-binding activity, and it is involved in the negative regulation of protein-binding activity, positive regulation of apoptotic process, and response to virus. In osteosarcoma cells, it is a cell death-promoting factor and can augment cisplatin-induced apoptosis (27), which is in line with our result. Similarly, lncRNA LINC00161 was reported to sensitize osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis (27). Indeed, interferon signaling is generally associated with tumor apoptosis or remission (28). A microarray study showed that the expression of IFIT2 is upregulated in ESCC (29). Therefore, it is reasonable to hypothesize that a high expression of IFIT2 plays a role in the anti-ESCC process. The role of BTN3A3 as a remission-associated mRNA in ESCC was another novel finding, and similar roles in tumor research have been reported. BTN3A3 is expected to become a potential therapeutic target for breast cancer (30), and the upregulation of BTN3A3 has been significantly correlated with better overall and relapse-free survival (31). Moreover, it has been noted to be a novel prognostic factor for hepatocellular carcinoma (32). In non-small cell lung cancer, knocking down BTN3A3 was shown to promote cell proliferation, migration, and invasion, and patients with a low expression of BTN3A3 have poor survival (33). A potential mechanism of its anticancer effect is that the BTN3A family proteins might be involved in the activation and proliferation of γδ T cells in the tumor microenvironment (34). Collectively, IFIT2, BTN3A, CCDC85B, SCD, and the hub genes in the remission/progression PPI networks (i.e., PSMB5, ZNF436, GTF2A2, COBLL1, AFP1, SOX5, MS4A1, CD79B, FUT7, CD3EAP, IMP4, DICER1, and ASH1L) are promising targets for ESCC treatment.
In this study, 4 miRNAs were found to be especially important: miR-484, miR-3615, miR-6803-3p, and miR-132-3p. Recently, miRNA expression profiling for the prediction of resistance to nCRT in ESCC has been reported (35), with miR-484 being reported to be an anti-ESCC marker. However, it is a carcinogenic miRNA, as shown in Figure 5H. Another study based on The Cancer Genome Atlas (TCGA) samples established a novel lncRNA panel for the prognosis of ESCC according to the competing endogenous RNA (ceRNA) mechanism (36). It was found that miR-3615 (another carcinogenic miRNA in our study) was positively correlated with patients’ overall survival. However, the association between miR-6803-3p or miR-132-3p and ESCC has not thus far been established. However, miR-132-3p does play an antitumoral role in bladder cancer, colorectal cancer, and osteosarcoma progression (37-39). In subsequent studies on ESCC, the roles of these 2 miRNAs warrant further attention.
This work has some limitations. First, we included only 15 patients and 3 groups, which may represent an insufficiently large sample to construct a prognostic model based on the correlations found in this study. Future studies will focus on the above key progression and remission markers in exo-RNAs and develop novel models to predict the treatment response to nCRT. Second, the lncRNA results showed no meaningful links to the miRNA-mRNA axes, either in the ceRNA networks or cis-regulatory networks. Our subsequent research will endeavor to clarify the molecular mechanisms, specifically, the miRNA precursor mechanism. Third, the difference in findings concerning the functions related to miR-484 and miR-3615 between our study and other works needs be further examined. Finally, in vitro and in vivo experiments are needed in the next research plan to validate the representative genes.
Conclusions
We identified the significantly altered plasma exo-RNAs among outcome groups and between different time points of nCRT, and clarified the corresponding enrichment and regulatory networks. They may serve as minimally invasive predictors of the progression and remission of patients with ESCC. In particular, several genes were found to have previously unknown roles in ESCC treatment, including IFIT2, BTN3A, CCDC85B, SCD, miR-484, miR-3615, miR-6803-3p, and miR-132-3p. The novel candidates in this study merit further investigation and may provide a deep understanding of the molecular mechanisms. Studies with a larger cohort of patients are needed to confirm the potential use of exosome transcriptome miRNAs as predictive biomarkers of the treatment response to nCRT.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (No. 2021YFC2501005) and the Shanghai Esophageal Cancer Cohort Database of Shanghai Hospital Development Center (No. SHDC2020CR6002).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-23-452/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-452/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-452/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-23-452/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institution ethics committee of Shanghai Chest Hospital (No. KS2160). All patients provided written informed consent for the use of blood and specimens.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Then EO, Lopez M, Saleem S, et al. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J Oncol 2020;11:55-64. [Crossref] [PubMed]
- Eyck BM, van Lanschot JJB, Hulshof MCCM, et al. Ten-Year Outcome of Neoadjuvant Chemoradiotherapy Plus Surgery for Esophageal Cancer: The Randomized Controlled CROSS Trial. J Clin Oncol 2021;39:1995-2004. [Crossref] [PubMed]
- Yang H, Liu H, Chen Y, et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. J Clin Oncol 2018;36:2796-803. [Crossref] [PubMed]
- Kilgour E, Rothwell DG, Brady G, et al. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020;37:485-95. [Crossref] [PubMed]
- Li L, Zhang L, Montgomery KC, et al. Advanced technologies for molecular diagnosis of cancer: State of pre-clinical tumor-derived exosome liquid biopsies. Mater Today Bio 2023;18:100538. [Crossref] [PubMed]
- Chen M, Xie Y, Luo Y, et al. Exosomes-a potential indicator and mediator of cleft lip and palate: a narrative review. Ann Transl Med 2021;9:1485. [Crossref] [PubMed]
- Wen X, Pu H, Liu Q, et al. Circulating tumor DNA-a novel biomarker of tumor progression and its favorable detection techniques. Cancers (Basel) 2022;14:6025. [Crossref] [PubMed]
- Han X, Zhou L, Tu Y, et al. Circulating exo-mir-154-5p regulates vascular dementia through endothelial progenitor cell-mediated angiogenesis. Front Cell Neurosci 2022;16:881175. [Crossref] [PubMed]
- Li K, Lin Y, Luo Y, et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer 2022;21:21. [Crossref] [PubMed]
- Todorova VK, Byrum SD, Gies AJ, et al. Circulating Exosomal microRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Curr Oncol 2022;29:613-30. [Crossref] [PubMed]
- Lei J, Zhang G, Li D, et al. Analysis of exosomal competing endogenous RNA network response to paclitaxel treatment reveals key genes in advanced gastric cancer. Front Oncol 2022;12:1027748. [Crossref] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013;310:2191-4. [Crossref] [PubMed]
- Meng F, Zhang X, Wang Y, et al. Hsa_circ_0021727 (circ-CD44) promotes ESCC progression by targeting miR-23b-5p to activate the TAB1/NFκB pathway. Cell Death Dis 2023;14:9. [Crossref] [PubMed]
- Fukuda T, Baba H, Okumura T, et al. miR-877-3p as a Potential Tumour Suppressor of Oesophageal Squamous Cell Carcinoma. Anticancer Res 2023;43:35-43. [Crossref] [PubMed]
- Shen Y, Shao Y, Ruan X, et al. Genetic variant in miR-17-92 cluster binding sites is associated with esophageal squamous cell carcinoma risk in Chinese population. BMC Cancer 2022;22:1253. [Crossref] [PubMed]
- Mohammadi E, Aliarab A, Babaei G, et al. MicroRNAs in esophageal squamous cell carcinoma: Application in prognosis, diagnosis, and drug delivery. Pathol Res Pract 2022;240:154196. [Crossref] [PubMed]
- Jiang L, Wang W, Li G, et al. High TUG1 expression is associated with chemotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. Cancer Chemother Pharmacol 2016;78:333-9. [Crossref] [PubMed]
- Wu J, Zhou Y, Wang T, et al. ZFX promotes tumorigenesis and confers chemotherapy resistance in esophageal squamous cell carcinoma. Clin Res Hepatol Gastroenterol 2021;45:101586. [Crossref] [PubMed]
- Lindner K, Eichelmann AK, Matuszcak C, et al. Complex Epigenetic Regulation of Chemotherapy Resistance and Biohlogy in Esophageal Squamous Cell Carcinoma via MicroRNAs. Int J Mol Sci 2018;19:499. [Crossref] [PubMed]
- Chang ZW, Jia YX, Zhang WJ, et al. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J Exp Clin Cancer Res 2018;37:56. [Crossref] [PubMed]
- Zhang Z, Xiong R, Li C, et al. LncRNA TUG1 promotes cisplatin resistance in esophageal squamous cell carcinoma cells by regulating Nrf2. Acta Biochim Biophys Sin (Shanghai) 2019;51:826-33. [Crossref] [PubMed]
- Wang J, Sahengbieke S, Xu X, et al. Gene expression analyses identify a relationship between stanniocalcin 2 and the malignant behavior of colorectal cancer. Onco Targets Ther 2018;11:7155-68. [Crossref] [PubMed]
- Du X, Wang Q, Hirohashi Y, et al. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp Mol Pathol 2006;81:184-90. [Crossref] [PubMed]
- Choi J, Gong JR, Hwang CY, et al. A Systems Biology Approach to Identifying a Master Regulator That Can Transform the Fast Growing Cellular State to a Slowly Growing One in Early Colorectal Cancer Development Model. Front Genet 2020;11:570546. [Crossref] [PubMed]
- Li LY, Yang Q, Jiang YY, et al. Interplay and cooperation between SREBF1 and master transcription factors regulate lipid metabolism and tumor-promoting pathways in squamous cancer. Nat Commun 2021;12:4362. [Crossref] [PubMed]
- Ma S, Zhou B, Yang Q, et al. A Transcriptional Regulatory Loop of Master Regulator Transcription Factors, PPARG, and Fatty Acid Synthesis Promotes Esophageal Adenocarcinoma. Cancer Res 2021;81:1216-29. [Crossref] [PubMed]
- Wang Y, Zhang L, Zheng X, et al. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett 2016;382:137-46. [Crossref] [PubMed]
- Dinh HQ, Pan F, Wang G, et al. Integrated single-cell transcriptome analysis reveals heterogeneity of esophageal squamous cell carcinoma microenvironment. Nat Commun 2021;12:7335. [Crossref] [PubMed]
- Chang HY, Lee CH, Li YS, et al. MicroRNA-146a suppresses tumor malignancy via targeting vimentin in esophageal squamous cell carcinoma cells with lower fibronectin membrane assembly. J Biomed Sci 2020;27:102. [Crossref] [PubMed]
- Chen S, Li Z, Huang W, et al. Prognostic and Therapeutic Significance of BTN3A Proteins in Tumors. J Cancer 2021;12:4505-12. [Crossref] [PubMed]
- Ren H, Li S, Liu X, et al. Multi-omics analysis of the expression and prognostic value of the butyrophilins in breast cancer. J Leukoc Biol 2021;110:1181-95. [Crossref] [PubMed]
- Huang C, Jiang X, Huang Y, et al. Identifying Dendritic Cell-Related Genes Through a Co-Expression Network to Construct a 12-Gene Risk-Scoring Model for Predicting Hepatocellular Carcinoma Prognosis. Front Mol Biosci 2021;8:636991. [Crossref] [PubMed]
- Cheng X, Ma T, Yi L, et al. Low expression of BTN3A3 indicates poor prognosis and promotes cell proliferation, migration and invasion in non-small cell lung cancer. Ann Transl Med 2021;9:478. [Crossref] [PubMed]
- Lu H, Dai W, Guo J, et al. High Abundance of Intratumoral γδ T Cells Favors a Better Prognosis in Head and Neck Squamous Cell Carcinoma: A Bioinformatic Analysis. Front Immunol 2020;11:573920. [Crossref] [PubMed]
- Slotta-Huspenina J, Drecoll E, Feith M, et al. MicroRNA expression profiling for the prediction of resistance to neoadjuvant radiochemotherapy in squamous cell carcinoma of the esophagus. J Transl Med 2018;16:109. [Crossref] [PubMed]
- Zhang J, Xiao F, Qiang G, et al. Novel lncRNA Panel as for Prognosis in Esophageal Squamous Cell Carcinoma Based on ceRNA Network Mechanism. Comput Math Methods Med 2021;2021:8020879. [Crossref] [PubMed]
- Li G, Guo BY, Wang HD, et al. CircRNA hsa_circ_0014130 function as a miR-132-3p sponge for playing oncogenic roles in bladder cancer via upregulating KCNJ12 expression. Cell Biol Toxicol 2022;38:1079-96. [Crossref] [PubMed]
- Guan H, Shang G, Cui Y, et al. Long noncoding RNA APTR contributes to osteosarcoma progression through repression of miR-132-3p and upregulation of yes-associated protein 1. J Cell Physiol 2019;234:8998-9007. [Crossref] [PubMed]
- He X, Ma J, Zhang M, et al. Long Non-Coding RNA SNHG16 Activates USP22 Expression to Promote Colorectal Cancer Progression by Sponging miR-132-3p. Onco Targets Ther 2020;13:4283-94. [Crossref] [PubMed]
(English Language Editor: J. Gray)


