
Circ_0003028 enhances the proliferation and glycolytic capacity and suppresses apoptosis in non-small cell lung cancer cells via the miR-1305/miR-1322-SLC5A1 axis
Highlight box
Key findings
• Circ_0003028 could accelerate the malignant behaviors and glycolytic capacity of NSCLC cells via a mechanism that may be related to the miR-1305 or miR-1322/SLC5A1 axis.
What is known and what is new?
• Circ_0003028 is located in chr14:66028054-66028484, the spliced length is 430 bp, the best transcript is NM_178155, and the gene symbol is FUT8. Circ_0003028 has also been mentioned in breast cancer.
• Circ_0003028 could accelerate the malignant behaviors and glycolytic capacity of NSCLC cells via a mechanism that may be related to miR-1305 or the miR-1322/SLC5A1 axis. Therefore, the findings of the current study provide a preliminary theoretical basis for NSCLC therapy and diagnosis.
What are the implications, and what should change now?
• We suggested that blocking the circ_0003028/miR-1305 or miR-1322/SLC5A1 axis might weaken the glycolytic pathway to achieve a therapeutic effect in NSCLC. However, this molecular axis still needs to be verified by extensive experiments in the future.
Introduction
Lung cancer is the most prevalent malignancy worldwide (1), especially non-small cell lung cancer (NSCLC) (2). At present, surgery remains the optimal treatment modality for early-stage NSCLC (3). However, for intermediate to advanced lung cancer, the currently available treatment options are still limited. After surgery, these intermediate and advanced patients may also have local recurrence or general metastasis (4). In recent years, new therapeutic approaches, such as immunotherapy and targeted therapy, have also increased the 5-year survival rate (5,6). However, these current therapeutic approaches are far from sufficient. Therefore, it is imperative to search for novel therapeutic targets, and exploring the mechanisms of NSCLC development is still extremely valuable.
Circular RNA (circRNA) is a novel endogenous non-coding RNA (ncRNA) that forms from exons or introns driven by a lasso or intron pairing (7). CircRNAs are abundant, highly conserved among species, and specifically expressed (8). They are also prominently resistant to RNA enzymes (7). The role of circRNA as a novel molecular marker in tumor diagnosis and treatment has attracted increasing attention. Studies have established that circRNA is abnormally expressed in tumor tissues and correlates with tumor progression (9,10). Hsa_circ_0000190 was directly involved in EGFR-MAPK-ERK signaling and might serve as a potential therapeutic target for the treatment of non-small cell lung carcinoma (11). Through literature analysis, we discovered that circ_0003028 was highly expressed in NSCLC (12), also differentially expressed circRNAs, including hsa_circ_0003028 had diagnostic value in NSCLC (13), and circ_0003028 also could enhance the proliferation, angiogenesis, and metastasis of NSCLC cells (14). According to the circBase database, circ_0003028 is located in chr14:66028054-66028484, the spliced length is 430 bp, the best transcript is NM_178155, and the gene symbol is fucosyltransferase 8 (FUT8). Moreover, circ_0003028 has also been mentioned in breast cancer (15). However, the influence of circ_0003028 on the apoptosis and glycolysis of NSCLC cells and the specific mechanism of circ_0003028 in NSCLC cells are not clear.
MicroRNAs (miRNAs), as small ncRNA molecules, can mediate different biological functions by binding mRNAs to play regulatory roles in genes (16,17). It has been reported that circRNA can function as a competitive endogenous RNA (ceRNA) or miRNA sponge (18). Also, the combination of miRNA and circRNA can form a minimal ceRNA network. CeRNAs can share miRNA recognition units, increasing the complexity of miRNA-based regulatory networks (19). Studies have reported that ceRNA networks exhibit significant potential in the diagnosis and targeted therapy of cancer (20,21). However, the function of circ_0003028/miRNA in NSCLC remains incompletely defined.
In the present study, we first confirmed the cyclic structure and stability of circ_0003028 and bioinformatics analysis showed that circ_0003028 might regulate glucose metabolism. We also verified the expression and prognosis of circ_0003028 in NSCLC. Furthermore, we investigated the impacts of circ_0003028 overexpression or silencing on the proliferation, apoptosis, and glycolysis of NSCLC cells. Moreover, we also explored the potential miRNAs associated with circ_0003028 and the possible target genes of the screened miRNAs. This study was conducted to provide potential targets for the therapy and diagnosis of NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-23-178/rc).
Methods
Clinical samples
A total of 104 NSCLC tissues and paired para-cancerous tissues were collected at the Sun Yat-sen University Cancer Center from August 2015 to June 2016. The inclusion criteria were as follows: (I) patients who had not received radiotherapy or chemotherapy before surgery; and (II) NSCLC was confirmed by postoperative pathological examination. The exclusion criteria were as follows: (I) incomplete medical records; (II) preoperative neoadjuvant therapy; (III) complicated with other systemic malignancies; and (IV) severe insufficiency of other organs. All tissues were rapidly stored at −80 ℃. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients who participated in this study signed an informed consent form, and this study was approved by the institutional ethics board of the Sun Yat-sen University Cancer Center (No. SL-B2022-643-01).
Cell culture
A549 and HCC827 cells were obtained from American Type Culture Collection (ATCC, Washington, DC, NW, USA). A549 cells and HCC827 cells were grown in RPMI-1640 (Gibco, C11875500BT). 10% fetal bovine serum (FBS, Gibco, USA) was added to both media, and the cells were cultured at 37 ℃ with 5% CO2.
Cell transfection
The empty vector, circ_0003028 overexpression plasmid, control short hairpin RNAs (shRNAs) (sh-CTRL), and circ_0003028 shRNAs (sh-circ) were obtained from HanBio Biotechnology (HanBio, Shanghai, China). MiRNA (miR)-1305 inhibitor, miR-1305 mimics, miR-1322 mimics, miR-1322 inhibitor, and negative control (NC) were acquired from GenePharma (Shanghai, China). The A549 and HCC827 cells (1×103 cells/well) were transfected with the above overexpressed plasmids and oligonucleotides by applying lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for 48 h based on the specification.
Agarose gel electrophoresis and sanger sequencing
Trizol (Invitrogen) was applied to isolate total cellular RNA, and a reverse transcription kit (R211-01, Vazyme Biotech, China) was utilized for reverse transcription. Genomic DNA (gDNA) was extracted from cells using a gDNA extraction kit (DP705, Tiangen Biochemical, China). Then, reverse transcription products [complementary DNA (cDNA) and gDNA] were amplified by SYBR Premix Ex TaqTM II (DRR081A, TaKaRa, Dalian, China). Divergent primers amplified the circular transcripts and convergent primers amplified the linear transcripts. Agarose gel electrophoresis was conducted and then placed on a gel imager system (E-GelTM Power Snap, Invitrogen, Carlsbad, CA, USA) for observation and photography. The circ_0003028 sequence was obtained from the circRNABase database. The polymerase chain reaction (PCR) amplification products were obtained by PCR and sent to Synbio Technologies (Suzhou, China) for sequencing.
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated from cells and milled tissues using Trizol (Invitrogen), and some of the extracted RNA was processed with ribonuclease R (RNaseR) (2 U/µg) at 37 ℃ for 10 min, and the control was processed with an equal amount of double-distilled water. After reverse transcription, the gene was amplified, and the data were analyzed with 2−ΔΔCT. Forward primer for circ_0003028 is 5'-GTCCAAGATTCTGGCAAAGC-3' and reverse primer for circ_0003028 is 5'-TCAAAGAGATCCTCCTGGTGA-3'. Forward primer for GAPDH is 5'-CTCCTCCTGTTCGACAGTCAGC-3' and reverse primer for GAPDH is 5'-CCCAATACGACCAAATCCGTT-3'.
Nucleoplasm separation experiment
Isolation of cytoplasm and nucleus from NSCLC cells was conducted using Cytoplasmic & Nuclear RNA Purification Kit (NGB-37400, NORGEN, ON, Canada), and circ_0003028 expression was analyzed via qRT-PCR in the nucleus and cytoplasm.
Cell proliferation
For cell counting kit-8 (CCK-8), NSCLC cells (4×105 cells/mL) in 96-well plates, and were transfected for 48 h based on the experimental purpose. Ten µL of CCK-8 (Dojindo, Tokyo, Japan) was supplemented to each well. After 2 h, the optical density (OD) value at 450 nm was tested. For 5-ethynyl-2'-deoxyuridine (EdU) staining, the processed cells were fixed with 4% paraformaldehyde, decolorized with 2 mg/mL glycine, and decolorized with 0.5% TritonX-100 for 10 min. Next, Apollo and DNA staining were conducted based on the instructions of the Cell-LightTM EdU Apollo567 In Vitro Kit (C10310-1, RIBOBIO, Guangzhou, China). After sealing the slices, the samples were photographed under a fluorescent microscope (Zeiss, Oberkochen, Germany).
Flow cytometer
The treated cells were harvested, washed using pre-chilled phosphate buffered saline (PBS), and resuspend using 500 µL of PBS. Subsequently, the cells were disposed of fluorescein isothiocyanate (FITC) (5 µL) and phycoerythrin (PE) (5 µL) for 10 min. The apoptotic cells were monitored by flow cytometry (FACS Calibur, BD, Franklin Lake, NJ, USA).
Determination of glucose, lactate, and adenosine triphosphate (ATP)
The levels of glucose consumption, lactate, and ATP synthesis in the A549 and HCC827 cells were monitored using glucose, lactate, and ATP kits (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions.
Detection of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR)
According to previous studies (22,23), the ECAR and OCR of NSCLC cells were determined using Seahorse XF Glycolysis or Cell Mito Stress Test kit (Seahorse Bioscience, Billerica, MA, USA) on the Seahorse XFe96 Extracellular Flux Analyzer.
Bioinformatics analysis
The target genes of miR-1322 and miR-1305 were predicted using DIANA-microT (https://mrmicrot.imsi.athenarc.gr/?r=mrmicrot/index) and TargetScan (https://www.targetscan.org/vert_80/).
Western blot
Total protein was harvested using RIPA lysate (Beyotime, China), and was monitored using a bicinchoninic acid (BCA) kit (Invitrogen). Proteins (40 µg) were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Boston, MA, USA). After blocking, the membranes were exposed to primary antibody (Abcam, Cambridge, England) overnight at 4 ℃, processed with secondary antibody (Abcam) for 1 h, and then developed by the dropwise addition of electrochemiluminescence (ECL) color development solution. After ECL chemiluminescence, the protein was developed on the gel imager (Bio-Rad, Hercules, CA, USA).
Statistical analysis
Measurement data were denoted as mean ± standard deviation (SD) from three experiments. SPSS22.0 software (SPSS, Inc., Chicago, IL, USA) was adopted for data analysis with the Student’s t-test or analysis of variance (ANOVA). A Kaplan-Meier survival curve was utilized to compare the overall survival between circ_0003028 and NSCLC patients. The prognosis was assessed using receiver operating characteristic (ROC) analysis. P<0.05 was considered statistically significant.
Results
High expression and poor prognosis of circ_0003028 in NSCLC
Firstly, we excluded genome rearrangement or trans-splicing. The PCR results showed that the divergent primers were amplified in cDNA but not in gDNA (Figure 1A). We then applied Sanger sequencing to the above PCR products, and the results displayed the head-to-tail splicing sites in the PCR products (Figure 1B). Meanwhile, we conducted qRT-PCR to evaluate the RNAs extracted from NSCLC cells treated with RNaseR, and the results demonstrated that circ_0003028 was resistant to RNaseR (Figure 1C). Next, we tested the circ_0003028 level by qRT-PCR and found that it was elevated in NSCLC tissues (Figure 1D). Based on the changes of circ_0003028 expression in NSCLC tissues, we also analyzed the relationship between circ_0003028 and the clinical characteristics of 104 NSCLC patients. We determined that the high expression of circ_0003028 was associated with the III–IV stage (P<0.0001), low differentiation (P=0.0016), and lymph node metastasis (P=0.0008) (Table 1).
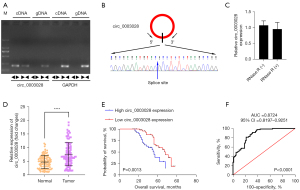
Table 1
| Factors | N | Hsa_circ_0003028 expression (n=104) | P value | |
|---|---|---|---|---|
| Low expression (n=52) | High expression (n=52) | |||
| Age (years) | 0.4314 | |||
| <60 | 48 | 26 | 22 | |
| ≥60 | 56 | 26 | 30 | |
| Gender | 0.4324 | |||
| Male | 50 | 23 | 27 | |
| Female | 54 | 29 | 25 | |
| TNM stage | <0.0001* | |||
| I–II | 46 | 33 | 13 | |
| III–IV | 58 | 19 | 39 | |
| Differentiation | 0.0016* | |||
| Poor | 56 | 20 | 36 | |
| Well | 48 | 32 | 16 | |
| Lymph node metastasis | 0.0008* | |||
| Yes | 57 | 20 | 37 | |
| No | 47 | 32 | 15 | |
| Smoking status | 0.2305 | |||
| Smoker | 62 | 28 | 34 | |
| Non-smoker | 42 | 24 | 18 | |
*P<0.05. NSCLC, non-small cell lung cancer; TNM, tumor-node-metastasis.
Subsequently, Kaplan-Meier analysis was performed to observe the probability of survival in NSCLC patients after recovery, and the schema showed that patients with a high expression of circ_0003028 had poor overall survival (Figure 1E). The ROC curve was utilized to determine the diagnostic potential of circ_0003028. As shown in the diagram, the results showed that the area under the curve (AUC) of circ_0003028 was 0.8724 [95% confidence interval (CI): 0.8197–0.9251], which suggested that circ_0003028 has a high diagnostic potential in NSCLC patients (Figure 1F).
Circ_0003028 induced proliferation and suppressed apoptosis in NSCLC cells
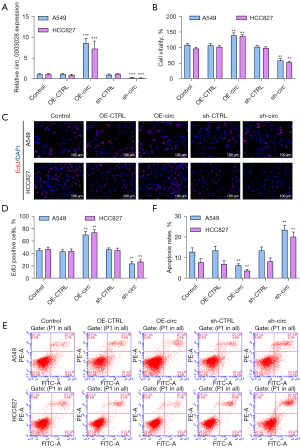
To explore the role of circ_0003028 in the development of NSCLC, we transfected A549 and HCC827 cells with circ_0003028 overexpressed plasmids or shRNAs, respectively. The circ_0003028 level in circ_0003028-overexpressed cells was markedly increased versus that in vector-transfected cells, and the opposite result trend was found in the circ_0003028-silenced cells (Figure 2A), which indicates that circ_0003028 was successfully overexpressed and silenced in A549 and HCC827 cells. Also, the CCK-8 and EdU results showed that the proliferation ability was dramatically enhanced in circ_0003028-overexpressed A549 and HCC827 cells compared to the overexpression-control group and was also markedly decreased in circ_0003028-silenced A549 and HCC827 cells relative to that in the sh-CTRL group (Figure 2B-2D). Meanwhile, the flow cytometer was used to monitor the cell apoptosis of the transfected cells, and the results demonstrated that cell apoptosis was conspicuously retarded in circ_0003028-overexpressed A549 and HCC827 cells. Circ_0003028 silencing induced cell apoptosis, which is consistent with the expected results (Figure 2E,2F).
Circ_0003028 overexpression elevated the glycolytic capacity of NSCLC cells
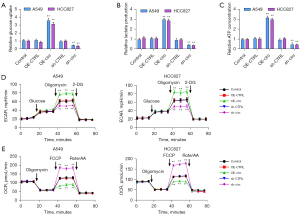
Further, we explored the function of circ_0003028 in the glycolysis of NSCLC cells. The results showed that circ_0003028 overexpression promoted glycolysis by increasing glucose uptake, lactate production, and ATP levels, while circ_0003028 down-regulation had the opposite effect (Figure 3A-3C). Moreover, the results demonstrated that circ_0003028 overexpression dramatically accelerated ECAR and diminished OCR, while circ_0003028 silencing prominently mitigated ECAR and strengthened OCR (Figure 3D,3E).
Circ_0003028 served as a sponge of miR-1305 and miR-1322
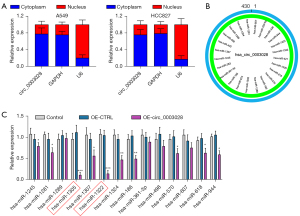
To confirm the subcellular distribution of circ_0003028, we observed the expression of cytoplasmic controlled transcripts glyceraldehyde-3-phosphate dehydrogenase (GAPDH), nuclear controlled transcripts U6, and circ_0003028 in the cytoplasm and nucleus of A549 and HCC827 cells, and the results revealed that circ_0003028 was mostly distributed in the cytoplasm (Figure 4A). We also screened the potential sponge miRNAs using the CircInteractome database (Figure 4B). Next, we selected miRNAs, and the results suggested that circ_0003028 overexpression substantially downregulated miR-1322, miR-1245, miR-1305, miR-1281, miR-186, miR-1289, miR-1307, miR-570, miR-1324, miR-618, and miR-944 in A549 cells, especially miR-1305 and miR-1322 (Figure 4C). Therefore, miR-1305 and miR-1322 were the most likely downstream regulators of circ_0003028.
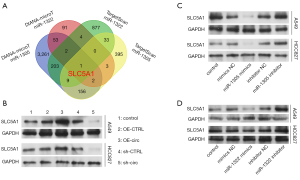
Solute carrier family 5 member 1 (SLC5A1) might be the target gene of miR-1305 and miR-1322
Based on the above results, we further identified the target genes of miR-1305 and miR-1322 that might play a key role in NSCLC. We first predicted the target genes by DIANA-microT and TargetScan to draw a Venn diagram of the cross-candidate target genes and found that SLC5A1 might be regulated by miR-1305 and miR-1322 (Figure 5A). Next, we verified whether SLC5A1 was the target gene of miR-1305 and miR-1322. The results showed that the SLC5A1 level was notably facilitated in circ_0003028-overexpressed NSCLC cells, and markedly retarded in circ_0003028-silenced NSCLC cells (Figure 5B). We then evaluated the SLC5A1 level in NSCLC cells after transfection with the miR-1305 mimics or inhibitor and miR-1322 mimics or inhibitor. The results revealed that the overexpression of miR-1305 or miR-1322 markedly decreased the SLC5A1 level, while inhibition of miR-1305 or miR-1322 prominently increased the SLC5A1 level in NSCLC cells (Figure 5C,5D).
Discussion
NSCLC is a malignant lung tumor (24), and patients with advanced NSCLC have a poor prognosis. In the era of precision cancer therapy, NSCLC treatment includes chemotherapy, immunotherapy, and targeted therapy (6,25). In the existing treatment model, the overall cure rate for NSCLC patients is <20%, and the 5-year survival rate for advanced patients is <5% (4). Therefore, it is crucial to determine an approach with good efficacy and few adverse effects to improve the survival rate and prolong the survival period of NSCLC patients.
CircRNA is a cyclic transcription sequence composed of exon or intron shears and is a naturally occurring endogenous ncRNA with a wide range and diversity (8,26). Studies have shown that circRNA is related to various tumors, including gastric cancer (27), pancreatic cancer (28), lung cancer (29), thyroid cancer (30), etc. Besides, circRNA has different biological properties and functions (31). There is a gradual increase in the number of reports of endogenous circRNAs associated with tumors (32,33). However, the functions of the vast majority of circRNAs have not yet been discovered. Previous reports have indicated that circ_0003028 expression is elevated in NSCLC (12), which could also accelerate the proliferation, angiogenesis, and metastasis of NSCLC cells (14). We further verified the circularity and stability of circ_0003028 and revealed the high expression of circ_0003028 in NSCLC tissues. Meanwhile, we discovered that the high expression of circ_0003028 can reduce the survival of NSCLC patients and has a certain prognostic effect. This suggests that circ_0003028 might have a catalytic effect on the NSCLC process.
Glucose metabolism is the main energy source for tumor cell growth and metabolism (34). The two primary methods of glucose oxidation in the body are oxidative phosphorylation and glycolysis. A previous study revealed that glycolysis can influence the biological behavior of tumor cells (35). The cell transports glucose into the cell via the glucose transporter (GLUT) on the membrane to produce pyruvate, a process known as glycolysis (36). Pyruvate is converted to lactate under hypoxic conditions and is metabolized by mitochondrial oxidative phosphorylation to produce energy under aerobic conditions. The active state of tumor cells is associated with the gained energy (37). Even when oxygen content is sufficient, tumor cells will also preferentially consume more glucose to produce energy through glycolysis (38).
Our results showed that circ_0003028 overexpression could increase glucose uptake, lactate production, and ATP concentration in NSCLC cells. Meanwhile, circ_0003028 overexpression could also elevate ECAR and reduce OCR in NSCLC cells. Furthermore, circ_0003028 silencing could exert the opposite effect to its overexpression. So, we confirmed that circ_0003028 could enhance the glycolysis of NSCLC cells. Our data also revealed that circ_0003028 could accelerate proliferation and weaken apoptosis in NSCLC cells. Overall, we proved that circ_0003028 has a significant acceleration effect on NSCLC progression.
By verifying the function of circ_0003028 in NSCLC cells, we further confirmed the possible mechanism of circ_0003028 in NSCLC. According to reports, circRNAs have miRNA response elements that bind to mRNAs and can thus contribute to the ability of miRNAs to regulate genes (39). As a molecular sponge of miRNA, circRNA can repress miRNA and affect the post-transcriptional regulation of target genes (18). In this way, we also screened the miRNAs that could be altered by circ_0003028 in NSCLC cells through bioinformatics and experimental verification. We also discovered that circ_0003028 might be a sponge of miR-1305 and miR-1322. Meanwhile, we further screened the common target genes of miR-1305 and miR-1322 through DIANA-microT and TargetScan. Through preliminary verification, we conclude that both miR-1305 and miR-1322 might regulate SLC5A1.
The SLC5A family are sodium-dependent GLUT proteins (SGLTs) (40). SGLTs have been reported to transport glucose into the intracellular SLC5A gene family independent of sodium ions (41). Among them, SGLT1 and SGLT2, which are encoded by SLC5A1 and SLC5A2, respectively, are GLUT proteins across the cell membrane (42). Specifically, SLC5A1 has been reported to exert a significant function in various cancer processes, including in pancreatic carcinoma (43), breast cancer (44,45), gastric cancer (46), endometrial cancer (47), etc. In our study, we further confirmed that circ_0003028 could upregulate SLC5A1, and miR-1305 or miR-1322 could downregulate SLC5A1. Therefore, we hypothesized that circ_0003028 could accelerate the NSCLC process via miR-1305 or the miR-1322/SLC5A1 axis to accelerate glycolysis.
For the shortcomings in the present study, first, a dual luciferase reporter system needs to be performed to confirm that circ_0003028 act as competing endogenous RNA (ceRNA) competitively bind to miR-1305 and miR-1322 to regulate SLC5A1 gene expression. Second, in vivo experiments need to be performed the determine the biological function of circ_0003028. Last, the problems and challenges in the clinical application of circRNAs, including improve the translation efficiency of circRNAs and overcome the off-target effect of circRNA knockdown technologies.
Conclusions
This study demonstrated that circ_0003028, as an oncogene, could accelerate the proliferation and glycolytic capacity and prevent the apoptosis of NSCLC cells. Also, the induction of glycolysis by circ_0003028 in NSCLC cells might occur via miR-1305 or the miR-1322/SLC5A1 axis. Therefore, we suggest that blocking the circ_0003028/miR-1305 or the miR-1322/SLC5A1 axis might weaken the glycolytic pathway to achieve a therapeutic effect in NSCLC, and glycolysis-associated multiomics prognostic model may provide potential application value for prognostic prediction and individualized treatment of NSCLC.
Acknowledgments
Funding: The research was supported by grants from the National Natural Science Foundation of China (Nos. 82274249 and 82074159), the Guangdong Basic and Applied Basic Research Foundation (Nos. 2021A1515011611 and 2021A1515010491), and the Guangdong Medical Science and Technology Research Fund (No. C2022062).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-23-178/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-178/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-23-178/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients who participated in this study signed an informed consent form, and this study was approved by the institutional ethics board of the Sun Yat-sen University Cancer Center (No. SL-B2022-643-01).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Mithoowani H, Febbraro M. Non-Small-Cell Lung Cancer in 2022: A Review for General Practitioners in Oncology. Curr Oncol 2022;29:1828-39. [Crossref] [PubMed]
- Nagano T, Tachihara M, Nishimura Y. Molecular Mechanisms and Targeted Therapies Including Immunotherapy for Non-Small Cell Lung Cancer. Curr Cancer Drug Targets 2019;19:595-630. [Crossref] [PubMed]
- Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 2019;27:167-70. [PubMed]
- Patel SA, Weiss J. Advances in the Treatment of Non-Small Cell Lung Cancer: Immunotherapy. Clin Chest Med 2020;41:237-47. [Crossref] [PubMed]
- Chiang CL, Huang HC, Luo YH, et al. Cerebrospinal fluid as a medium of liquid biopsy in the management of patients with non-small-cell lung cancer having central nervous system metastasis. Front Biosci (Landmark Ed) 2021;26:1679-88. [Crossref] [PubMed]
- Zhou M, Xiao MS, Li Z, et al. New progresses of circular RNA biology: from nuclear export to degradation. RNA Biol 2021;18:1365-73. [Crossref] [PubMed]
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 2020;19:172. [Crossref] [PubMed]
- Sun W, Zhou H, Han X, et al. Circular RNA: A novel type of biomarker for glioma Mol Med Rep 2021;24:602. (Review). [Crossref] [PubMed]
- Zhang S, Sun J, Gu M, et al. Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Med 2021;10:8725-40. [Crossref] [PubMed]
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 Promotes Non-Small Cell Lung Cancer via Modulation of the EGFR/ERK Pathway. Cancer Res 2022;82:75-89. [Crossref] [PubMed]
- Zhou Y, Zhan Y, Jiang W, et al. Long Noncoding RNAs and Circular RNAs in the Metabolic Reprogramming of Lung Cancer: Functions, Mechanisms, and Clinical Potential. Oxid Med Cell Longev 2022;2022:4802338. [Crossref] [PubMed]
- Li L, Sun D, Li X, et al. Identification of Key circRNAs in Non-Small Cell Lung Cancer. Am J Med Sci 2021;361:98-105. [Crossref] [PubMed]
- Guan H, Sun C, Gu Y, et al. Circular RNA circ_0003028 contributes to tumorigenesis by regulating GOT2 via miR-1298-5p in non-small cell lung cancer. Bioengineered 2021;12:2326-40. [Crossref] [PubMed]
- Firoozi Z, Mansoori Y, Saeidi K, et al. Experimental and Bioinformatic Clues to the Potential Roles of hsa_circ_0013958 and hsa_circ_0003028 in Clinopathophysiology of Breast Cancer. Galen Medical Journal 2021;10:e2064. [Crossref]
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019;234:5451-65. [Crossref] [PubMed]
- Banerjee S, Thompson WE, Chowdhury I. Emerging roles of microRNAs in the regulation of Toll-like receptor (TLR)-signaling. Front Biosci (Landmark Ed) 2021;26:771-96. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ 2022;29:481-91. [Crossref] [PubMed]
- Li J, Sun D, Pu W, et al. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020;6:319-36. [Crossref] [PubMed]
- Zhang M, Bai X, Zeng X, et al. circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta 2021;523:120-30. [Crossref] [PubMed]
- Zhang Y, Wu Y, Su X. PLOD1 promotes cell growth and aerobic glycolysis by regulating the SOX9/PI3K/Akt/mTOR signaling pathway in gastric cancer. Front Biosci (Landmark Ed) 2021;26:322-34. [Crossref] [PubMed]
- Ozsvari B, Nuttall JR, Sotgia F, et al. Azithromycin and Roxithromycin define a new family of "senolytic" drugs that target senescent human fibroblasts. Aging (Albany NY) 2018;10:3294-307. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Chen L, Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett 2021;505:49-57. [Crossref] [PubMed]
- Li R, Jiang J, Shi H, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci 2020;77:1661-80. [Crossref] [PubMed]
- Rong Z, Xu J, Shi S, et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics 2021;11:2755-69. [Crossref] [PubMed]
- Wang C, Tan S, Li J, et al. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett 2020;492:106-15. [Crossref] [PubMed]
- Zhu G, Chang X, Kang Y, et al. CircRNA: A novel potential strategy to treat thyroid cancer Int J Mol Med 2021;48:201. (Review). [Crossref] [PubMed]
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 2019;12:90. [Crossref] [PubMed]
- Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 2022;19:188-206. [Crossref] [PubMed]
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer 2020;19:30. [Crossref] [PubMed]
- Mulukutla BC, Yongky A, Le T, et al. Regulation of Glucose Metabolism - A Perspective From Cell Bioprocessing. Trends Biotechnol 2016;34:638-51. [Crossref] [PubMed]
- Bose S, Le A. Glucose Metabolism in Cancer. Adv Exp Med Biol 2018;1063:3-12. [Crossref] [PubMed]
- Lu YY, Wu CH, Hong CH, et al. GLUT-1 Enhances Glycolysis, Oxidative Stress, and Fibroblast Proliferation in Keloid. Life (Basel) 2021;11:505. [Crossref] [PubMed]
- Bergers G, Fendt SM. The metabolism of cancer cells during metastasis. Nat Rev Cancer 2021;21:162-80. [Crossref] [PubMed]
- Schwartz L, Supuran CT, Alfarouk KO. The Warburg Effect and the Hallmarks of Cancer. Anticancer Agents Med Chem 2017;17:164-70. [Crossref] [PubMed]
- Alkan AH, Akgül B. Endogenous miRNA Sponges. Methods Mol Biol 2022;2257:91-104. [Crossref] [PubMed]
- Chiba Y, Murakami R, Matsumoto K, et al. Glucose, Fructose, and Urate Transporters in the Choroid Plexus Epithelium. Int J Mol Sci 2020;21:7230. [Crossref] [PubMed]
- Harada N, Inagaki N. Role of sodium-glucose transporters in glucose uptake of the intestine and kidney. J Diabetes Investig 2012;3:352-3. [Crossref] [PubMed]
- Rieg T, Vallon V. Development of SGLT1 and SGLT2 inhibitors. Diabetologia 2018;61:2079-86. [Crossref] [PubMed]
- Gao HF, Chen LY, Cheng CS, et al. SLC5A1 promotes growth and proliferation of pancreatic carcinoma via glucose-dependent AMPK/mTOR signaling. Cancer Manag Res 2019;11:3171-85. [Crossref] [PubMed]
- Liu H, Ertay A, Peng P, et al. SGLT1 is required for the survival of triple-negative breast cancer cells via potentiation of EGFR activity. Mol Oncol 2019;13:1874-86. [Crossref] [PubMed]
- Wang J, Ji H, Niu X, et al. Sodium-Dependent Glucose Transporter 1 (SGLT1) Stabled by HER2 Promotes Breast Cancer Cell Proliferation by Activation of the PI3K/Akt/mTOR Signaling Pathway in HER2+ Breast Cancer. Dis Markers 2020;2020:6103542. [Crossref] [PubMed]
- Shi M, Wang C, Ji J, et al. CRISPR/Cas9-mediated knockout of SGLT1 inhibits proliferation and alters metabolism of gastric cancer cells. Cell Signal 2022;90:110192. [Crossref] [PubMed]
- Zeng N, Okumura T, Alauddin M, et al. LEFTY2/endometrial bleeding-associated factor up-regulates Na+ Coupled Glucose Transporter SGLT1 expression and Glycogen Accumulation in Endometrial Cancer Cells. PLoS One 2020;15:e0230044. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


