
Clinical relevance and distribution of Helicobacter pylori virulence factors in isolates from Chinese patients
Highlight box
Key findings
• The virulence factors including cagA, cagE, vacA s1, jhp0562, homB and hopQI were widely distributed in our study. There was a strong correlation between the hrgA gene and gastric carcinoma.
What is known and what is new?
• H. pylori is characterized by genetic diversity, and virulence factors present different patterns of distribution within clinical strains. Studies have revealed CagA is considered as an important carcinogen, and the genotype vacA s1m1 is highly related with ulcers or gastric cancer.
• This study investigated the distribution of cagA, vacA, jhp0562, jhp0563, homA, homB, hopQI, hopQII, hrgA, and hpyIIIR in H. pylori isolates from patients with different gastrointestinal diseases assessed their association with clinical outcomes.
What is the implication, and what should change now?
• The study indicated that hrgA may be a potential independent factor associated with severe diseases and may predict the future development of GC in such patients.
Introduction
Helicobacter pylori (H. pylori) infections can cause severe gastroduodenal diseases, including chronic gastritis (CG), peptic ulcer disease (PUD), gastric carcinoma (GC), and mucosa-associated lymphoid tissue lymphoma (1). It is reported that majority of H. pylori-infected individuals remain asymptomatic throughout life and some individuals have the risk of developing severe disease, such as GC. The bacterial factors have been implicated in various clinical symptoms, especially numerous virulence factors with heterogeneity across strains (2,3). Various virulence factors enable the survival of H. pylori in the acidic environment, successful colonization of the gastric mucosa, and persistent infection, causing chronic inflammation and tissue damage (3). The most well-studied factor is cytotoxin-associated gene A (cagA), which has been considered an important carcinogen (4). In addition, several other virulence factors may also play an important role in the pathogenicity of H. pylori (5).
CagE, which is the second most studied gene of the cag pathogenicity island (cagPAI), is essential for CagA translocation and phosphorylation (6). Studies have revealed that it is a better marker for cagPAI presence than cagA and is considered an important risk factor for duodenal ulcer (DU) development (7,8). VacA is present in almost all H. pylori strains, and the different combinations of s and m regions determine the cytotoxic activity of different strains (9). Previous studies showed that the genotype s1m1 is highly related with PUD or GC (10,11). Jhp0562, which is involved in the synthesis of lipopolysaccharide, is located upstream of the jhp0563 gene (12). The jhp0562 and jhp0563 genes are highly similar (>80%) and are highly associated with PUD (13-15). Many studies have focused on the homA and homB genes, which are 90% identical (16,17). Previous studies showed that the presence of homB was associated with PUD and development of GC (18,19). HopQ belongs to the largest outer membrane protein family in H. pylori, and two families of hopQ alleles have been described, namely, hopQI and hopQII (20,21). The hopQI genotype has been commonly detected in both Western and East Asian strains, whereas the hopQII genotype is prevalent in Western strains (22). Additionally, a previous report revealed a correlation between the hopQI gene and PUD (23). In numerous Asian and Western strains, a novel hrgA gene has been identified upstream of hpyIIIM in the hpyIII R-M system of H. pylori (24,25). Previous studies suggested that the hrgA gene frequency increased among GC patients and indicated that hrgA might be a marker for GC development among East Asian patients (26,27).
Due to geographical variations, the distribution and roles of these virulence factors in gastric diseases are still under investigation. Furthermore, there is a paucity of data regarded the distribution of cagE, jhp0562, jhp0563, and hrgA in China, which has a higher H. pylori infection incidence and GC occurrence than Western countries. Heilongjiang (HLJ) and Jiangxi (JX), with significant geographic variations, are high-risk areas for the development of GC. Therefore, we investigated the distribution of the cagA, cagE, vacA, jhp0562, jhp0563, homA, homB, hopQI, hopQII, hrgA, and hpyIIIR genes in clinical strains isolated from patients with gastroduodenal diseases in HLJ and JX to assess the association between diseases and virulence factors. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-23-1404/rc).
Methods
Study subjects
A total of 160 patients were involved in this study, including 59 patients from the Affiliated Hospital of Harbin Medical University (Heilongjiang Province, China) and 101 patients from the First Affiliated Hospital of Nanchang University (Jiangxi Province, China). The gastrointestinal disease type was determined by gastrointestinal endoscopy and histopathological tests. The gastric mucosal biopsies were taken from the greater curvature of the gastric antrum during upper gastrointestinal endoscopy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention (Approval No. ICDC-2013001) and written informed consent was obtained from all patients.
Culture and extraction of genomic DNA
Gastric biopsy specimens were homogenized thoroughly and streaked onto Karmali agar plates (Oxoid) supplemented with 5% fresh defibrinated sheep blood and cultured under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) at 37 ℃ for 48 hours. H. pylori colonies were identified by typical morphology, Gram staining, and urease, oxidase, and catalase activity tests. The bacterial cells were washed twice with phosphate buffered saline (PBS) and centrifuged at 8,000 rpm for 3 minutes. The genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions.
Polymerase chain reaction (PCR) amplification
The PCR reaction was performed in a volume of 25 µL, containing forward and reverse primers (1 µM). The amplification was as follows: 94 ℃ for 5 minutes, followed by 35 cycles of 94 ℃ for 45 seconds, 54 ℃ for 45 seconds, and 72 ℃ for 45 seconds, and finally 72 ℃ for 5 minutes. The primers used to amplify the targeted genes are summarized in Table 1. The H. pylori strain 26695 and strain J99 were used as controls. The PCR products were analyzed on a 1% agarose gel containing 1 X TAE (Tris, acetic acid, EDTA) buffer.
Table 1
| Primer name | Primer sequence (5'-3') | Product size | Reference |
|---|---|---|---|
| cagA | F-TGCGTGTGTGGCTGTTAGTAG | 593 bp | (28) |
| R-CCTAGTCGGTAATGGGTTGT | |||
| cagE | F-TTGAAAACTTCAAGGATAGGATAGAGC | 508 bp | (29) |
| R-GCCTAGCGTAATATCACCATTACCC | |||
| vacA s1/s2 | F-ATGGAAATACAACAAACACAC | 259 bp/286 bp | (28) |
| R-CTGCTTGAATGCGCCAAAC | |||
| vacA m1 | F-GGCCCCAATGCAGTCATGGAT | 240 bp | (28) |
| R-GCTGTTAGTGCCTAAAGAAGCAT | |||
| vacA m2 | F-GGAGCCCCAGGAAACATTG | 352 bp | (28) |
| R-CATAACTAGCGCCTTGCAC | |||
| jhp0562/jhp0563 | F-TGAAAAGCCCTTTTGATTTTG | 301 bp/602 bp | (5) |
| R-GCTGTAGTGGCCACATACACG | |||
| homA/homB | F-AGAGGGTGTTTGAAACGCTCAATA | 128 bp/161 bp | (14) |
| R-GGTGAATTCTTCTGCGGTTTG | |||
| hopQI | F-ACG AACGCGCAAAAACTTTA | 187 bp | (30) |
| R-TTGCCATTCTCATCGGTGTA | |||
| hopQII | F-ACAGCCACTCCAATCCAGAA | 160 bp | (30) |
| R-AACCCCACCGTGGATTTTAG | |||
| hrgA | F-TCTCGTGAAAGAGAATTTCC | 594 bp | (31) |
| R-TAAGTGTGGGTATATCAATC | |||
| hpyIIIR | F-CTCATTGCTGTGAGGGAT | 420 bp | (31) |
| R-TCTTGATAGGATCTTGCG |
The primer sequences used for the genes were the same as the references. F, forward primer sequence; R, reverse primer sequence.
Statistical analysis
The relationship between each genotype and different regions, as well as clinical outcomes, was quantified using the chi-squared test. A P value (bilateral test) less than 0.05 was considered statistically significant. The logistic models were used to explore the relationship between diseases and the candidate genes. The odds ratio and 95% confidence interval (CI) were obtained for multivariate analysis. When one condition (e.g., GC) was studied, the other conditions were used as reference (e.g., CG and PUD). Initially, the full model was used, which included ‘age’, ‘sex’, and all candidate genes. Thereafter, the model was subjected to backward stepwise selection for candidate risk factors. The R function ‘glm ()’ was used to build the full model and the ‘MASS: stepAIC ()’ function was used to perform model selection. The R function ‘rcorr ()’ was used to calculate the Spearman correlation coefficient between studied genes.
Results
A total of 160 H. pylori strains were successfully isolated from gastric biopsy specimens, including 59 from HLJ (39 with CG, 9 with PUD, and 11 with GC) and 101 from JX (38 with CG, 36 with PUD, and 27 with GC). There was no significant difference in the sex nor age of the patients between the two regions. The average age of the examined patients was 47.5±15.2 years, and the average age in patients with GC was significantly higher than that in CG and PUD patients. The percentage of male patients with GC was significantly higher than that of CG patients (P<0.05; Table 2).
Table 2
| Characteristics | HLJ [59] | JX [101] | Total [160] | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | PUD | GC | CG | PUD | GC | CG | PUD | GC | |||
| n | 39 | 9 | 11 | 38 | 36 | 27 | 77 | 45 | 38 | ||
| Mean age ± SD (years) | 46.6±10.5 | 46.1±13.5 | 64.9±10.9 | 41.6±12.1 | 41.9±12.8 | 57.3±12 | 43.3±12.2* | 42.8±113 | 59.5±12.2 | ||
| Male | 17 (43.6%) | 6 (66.7%) | 9 (81.5%) | 20 (52.6%) | 23 (63.9%) | 17 (62.9%) | 37 (48.1%)* | 29 (64.4%) | 26 (68.4%)* | ||
| Total mean age ± SD (years) | 51.5±14.43 | 45.9±14.12 | 47.5±15.2 | ||||||||
| Total male | 32 (54.2%) | 60 (59.4%) | 92 (57.5%) | ||||||||
*, indicate a significant difference. CG, chronic gastritis; PUD, peptic ulcer disease; GC, gastric carcinoma; JX, Jiangxi; HLJ, Heilongjiang.
The presence of virulence factors in all 160 strains was determined by PCR and the results are summarized in Table 3.
Table 3
| Genotypes | No. of isolates | ||
|---|---|---|---|
| HLJ (n=59) | JX (n=101) | Total (n=160) | |
| cagA | 59 (100.0) | 101 (100.0) | 160 (100.0) |
| cagE | 59 (100.0) | 101 (100.0) | 160 (100.0) |
| vacA | |||
| s1 | 58 (98.3) | 100 (99.0) | 158 (98.8) |
| s2 | 1 (1.7) | 1 (1.0) | 2 (1.2) |
| m1 | 18 (30.5) | 33 (32.7) | 51 (31.9) |
| m2 | 41 (69.5) | 68 (67.3) | 109 (68.1) |
| s1m1 | 18 (30.5) | 32 (31.7) | 50 (31.3) |
| s1m2 | 40 (67.8) | 68 (67.3) | 108 (67.5) |
| s2m1 | 0 | 1 (1.0) | 1 (0.6) |
| s2m2 | 1 (1.7) | 0 | 1 (0.6) |
| jhp0562/ | |||
| jhp0562+ | 58 (97.5) | 101 (100.0) | 159 (99.4) |
| jhp0563 | |||
| jhp0563+ | 19 (32.2) | 33 (32.7) | 52 (32.5) |
| jhp0562+/jhp0563- | 40 (67.8) | 68 (67.3) | 108 (67.5) |
| jhp0562-/jhp0563+ | 1 (1.7) | 0 | 1 (0.6) |
| Double positive | 18 (30.5) | 33 (32.7) | 51 (31.9) |
| homA/ | |||
| homA+ | 20 (33.9) | 33 (32.7) | 53 (33.1) |
| homB | |||
| homB+ | 41 (69.5) | 73 (72.3) | 114 (71.3) |
| homA+/homB- | 18 (30.5) | 28 (27.7) | 46 (28.8) |
| homA-/homB+ | 39 (66.1) | 68 (67.3) | 107 (66.9) |
| Double positive | 2 (3.4) | 5 (5.0) | 7 (4.4) |
| hopQI/ | |||
| hopQ I+ | 59 (100.0) | 101 (100.0) | 160 (100.0) |
| hopQII | |||
| hopQ II+ | 8 (13.6)* | 3 (3.0)* | 11 (6.9) |
| hopQ I+/hopQ II- | 51 (86.4)* | 98 (97.0)* | 149 (93.1) |
| hopQ I-/hopQ II+ | 0 | 0 | 0 |
| Double positive | 8 (13.6)* | 3 (3.0)* | 11 (7.0) |
| hrgA/ | |||
| hrgA+ | 31 (52.5) | 60 (59.4) | 91 (56.9) |
| hpyIIIR | |||
| hpyIIIR+ | 45 (76.3) | 88 (87.1) | 133 (83.1) |
| hrgA+/hpyIIIR- | 14 (23.7)* | 11 (10.9)* | 25 (15.6) |
| hrgA-/hpyIIIR+ | 28 (47.5) | 39 (38.6) | 67 (41.9) |
| Double positive | 17 (28.8)* | 49 (48.5)* | 66 (41.3) |
| None | 0 | 2 (2.0) | 2 (1.3) |
*, indicate a significant difference. +, the positive genotype; −, the negative genotype; HLJ, Heilongjiang; JX, Jiangxi; n, the number of strains.
cagA, cagE, and vacA status
All strains tested were successfully detected for the cagA and cagE genes. The most common vacA s genotype was s1 (158/98.8%), while the vacA s2 subtype was detected in only two (1.2%) strains in our study (Table 3, Figure 1). For the vacA m region, the m2 subtype was present in 109 (68.1%) strains, and the m1 subtype was present in 51 (31.9%) strains. The frequencies of the m1 subtype strains were 30.5% and 32.7% in HLJ and JX, respectively. Conversely, the m2 subtype was present in 69.5% and 67.3% of strains from HLJ and JX, respectively (Table 3, Figure 1A,1B). There were no significant differences in the m subtypes between different geographic regions (P>0.05). The dominant vacA combination was s1m2 (67.5%), which was significantly higher than s1m1 (31.3%, P<0.001), but no statistical significance was obtained between the different regions (P>0.05; Table 3).

The dominant m2 subtype was detected in 71.1%, 73.7%, and 63.6% of strains isolated from patients with PUD, GC, and CG, respectively. However, the difference was not significant (P>0.05). In the present study, the vacA s2 subtype was present in only two strains isolated from CG patients, and there were no patients with PUD and GC infected with s2 subtype strains (Table 4, Figure 1C-1E).
Table 4
| Genotypes | No. of isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HLJ (n=59) | JX (n=101) | Total (n=160) | |||||||||
| CG [39] | PUD [9] | GC [11] | CG [38] | PUD [36] | GC [27] | CG [77] | PUD [45] | GC [38] | |||
| cagA | 39 (100.0) | 9 (100.0) | 11 (100.0) | 38 (100.0) | 36 (100.0) | 27 (100.0) | 77 (100.0) | 45 (100.0) | 38 (100.0) | ||
| cagE | 39 (100.0) | 9 (100.0) | 11 (100.0) | 38 (100.0) | 36 (100.0) | 27 (100.0) | 77 (100.0) | 45 (100.0) | 38 (100.0) | ||
| vacA | |||||||||||
| s1 | 38 (97.4) | 9 (100.0) | 11 (100.0) | 37 (97.4) | 36 (100.0) | 27 (100.0) | 75 (97.4) | 45 (100.0) | 38 (100.0) | ||
| s2 | 1 (2.6) | 0 | 0 | 1 (2.6) | 0 | 0 | 2 (2.6) | 0 | 0 | ||
| m1 | 15 (38.5) | 1 (11.1) | 2 (18.2) | 13 (34.2) | 12 (33.3) | 8 (29.6) | 28 (36.4) | 13 (28.9) | 10 (26.3) | ||
| m2 | 24 (61.5) | 8 (88.9) | 9 (81.8) | 25 (65.8) | 24 (66.7) | 19 (70.4) | 49 (63.6) | 32 (71.1) | 28 (73.7) | ||
| s1m1 | 15 (38.5) | 1 (11.1) | 2 (18.2) | 12 (31.6) | 12 (33.3) | 8 (29.6) | 27 (35.1) | 13 (28.9) | 10 (26.3) | ||
| s1m2 | 23 (58.9) | 8 (88.9) | 9 (81.8) | 25 (65.8) | 24 (66.7) | 19 (70.4) | 48 (62.3) | 32 (71.1) | 28 (73.7) | ||
| s2m1 | 0 | 0 | 0 | 1 (2.6) | 1 (1.3) | 0 | 0 | ||||
| s2m2 | 1 (2.6) | 0 | 0 | 0 | 0 | 0 | 1 (1.3) | 0 | 0 | ||
| jhp0562+ | 39 (100.0) | 9 (100.0) | 10 (90.9) | 38 (100.0) | 36 (100.0) | 27 (100.0) | 77 (100.0) | 45 (100.0) | 37 (97.4) | ||
| jhp0563+ | 13 (33.3) | 4 (44.4) | 2 (18.2) | 13 (34.2) | 13 (36.1) | 7 (25.9) | 26 (33.8) | 17 (37.8) | 9 (23.7) | ||
| jhp0562+/jhp0563- | 26 (66.7) | 5 (55.6) | 9 (81.8) | 25 (65.8) | 23 (63.9) | 20 (74.1) | 51 (66.2) | 28 (62.2) | 29 (76.3) | ||
| jhp0562-/jhp0563+ | 0 | 0 | 1 (9.1) | 0 | 0 | 0 | 0 | 0 | 1 (2.63) | ||
| jhp0562+/jhp0563+ | 13 (33.3) | 4 (44.4) | 1 (9.1) | 13 (34.2) | 13 (36.1) | 7 (25.9) | 26 (33.8) | 17 (37.8) | 8 (21.1) | ||
| homA+ | 15 (38.5) | 1 (11.1) | 4 (36.4) | 12 (31.6) | 12 (33.3) | 9 (33.3) | 27 (35.1) | 13 (28.9) | 13 (34.2) | ||
| homB+ | 26 (66.7) | 8 (88.9) | 7 (63.6) | 29 (76.3) | 26 (72.2) | 18 (66.7) | 55 (71.4) | 34 (75.6) | 25 (65.8) | ||
| homA+/homB- | 13 (33.3) | 1 (11.1) | 4 (36.4) | 9 (23.7) | 10 (27.8) | 9 (33.3) | 22 (28.6) | 11 (24.4) | 13 (34.2) | ||
| homA-/homB+ | 24 (61.5) | 8 (88.9) | 7 (63.6) | 26 (68.4) | 24 (66.7) | 18 (66.7) | 50 (64.9) | 32 (71.1) | 25 (65.8) | ||
| homA+/homB+ | 2 (5.1) | 0 | 0 | 3 (7.9) | 2 (5.6) | 0 | 5 (6.5) | 2 (4.4) | 0 | ||
| hopQ I+ | 39 (100.0) | 9 (100.0) | 11 (100.0) | 38 (100.0) | 36 (100.0) | 27 (100.0) | 77 (100.0) | 45 (100.0) | 38 (100.0) | ||
| hopQ II+ | 5 (12.8) | 1 (11.1) | 2 (18.2) | 0 | 2 (5.6) | 1 (3.7) | 5 (6.5) | 3 (6.7) | 3 (7.9) | ||
| hopQ I+/hopQ II- | 34 (87.2) | 8 (88.9) | 9 (81.8) | 38 (100.0) | 34 (94.4) | 26 (96.3) | 72 (93.5) | 42 (93.3) | 35 (92.1) | ||
| hopQ I-/hopQ II+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| hopQ I+/hopQ II+ | 5 (12.8) | 1 (11.1) | 2 (18.2) | 0 | 2 (5.6) | 1 (3.7) | 5 (6.5) | 3 (6.7) | 3 (7.9) | ||
| hrgA+ | 21 (53.8) | 4 (44.4) | 6 (54.5) | 18 (46.4)* | 21 (58.3) | 21 (77.8)* | 39 (50.7)* | 25 (55.6) | 27 (71.1)* | ||
| hpyIIIR+ | 31 (79.5) | 7 (77.8) | 7 (63.6)* | 31 (81.6) | 32 (88.9) | 25 (92.6)* | 62 (80.5) | 39 (86.7) | 32 (84.2) | ||
| hrgA+/hpyIIIR- | 8 (20.5) | 2 (22.2) | 4 (36.4)* | 7 (18.4) | 2 (5.6) | 2 (7.4)* | 15 (19.5) | 4 (8.9) | 6 (15.8) | ||
| hrgA-/hpyIIIR+ | 18 (46.2) | 5 (55.6) | 5 (45.4) | 20 (52.6)* | 13 (36.1) | 6 (22.2)* | 38 (49.4)* | 18 (40.0) | 11 (28.9)* | ||
| hrgA+/hpyIIIR + | 13 (33.3) | 2 (22.2) | 2 (18.2)* | 11 (28.9)* | 19 (52.8) | 19 (70.4)* | 24 (31.2)* | 21 (46.7) | 21 (55.3)* | ||
| None | 0 | 0 | 0 | 0 | 2 (5.6) | 0 | 0 | 2 (4.4) | 0 | ||
Values in parentheses are percentages. *, indicate a significant difference. +, the positive genotype; −, the negative genotype; HLJ, Heilongjiang; JX, Jiangxi; CG, chronic gastritis; PUD, peptic ulcer disease; GC, gastric carcinoma; n, the number of strains.
jhp0562 and jhp0563 status
In this study, three different combinations of jhp0562 and jhp0563 were observed: a single fragment of jhp0562 (profile 1) or jhp0563 (profile 2), and a double positive for two genes (profile 3) with different lengths of jhp0563 (Figure 2A).
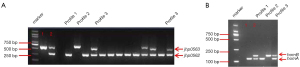
Overall, 99.4% of the strains carried the jhp0562 gene in our study, which was much higher than those carrying jhp0563 (32.5%, P<0.001). The dominant jhp0562-single positive genotype was detected in 108 (67.5%) strains, whereas the jhp0563-single positive genotype was observed in only 1 strain. There was no significant difference in the prevalence of any of the genotypes between HLJ and JX (Table 3).
Jhp0563 was present in 33.8% of isolates from CG patients and 37.8% from PUD patients, which was higher than that from GC patients (23.7%), however, the difference was not statistically significant (P>0.05). No correlation between jhp0562 single-positive genotype and clinical outcome was observed (P>0.05). The prevalence of double positive strains in patients with CG, PUD, and GC was 33.8%, 37.8%, and 21.1%, respectively. There was also no significant difference between the mixed genotype and various diseases (P>0.05; Table 4).
homA and homB status
The clinical strains can have a single homA gene (profile 1), a single homB gene (profile 2), or both homA and homB genes (profile 3) (Figure 2B).
The homA gene was detected in 53 (33.1%) strains, whereas the homB gene (71.3%) was more frequently detected than homA (P<0.001). Moreover, double positivity for homA and homB strains was rare (4.4%). The homB-single positive genotype was dominant, accounting for 66.9% of strains, whereas the homA-single positive genotype was present in 28.8% of the strains. We found no significant difference in the prevalence of homA or homB between the two geographical regions (Table 3).
A total of 71.4% of CG patients and 75.6% of PUD patients were infected with homB-positive strains, which were higher than the percentage of GC patients (65.8%). However, the difference was not statistically significant (P>0.05). homB single-positive strains were present in 64.9% of CG patients, 71.1% of PUD patients, and 65.8% of GC patients, whereas strains with homA single-positive genotypes accounted for 28.6%, 24.4%, and 34.2%, respectively. In this study, there was no association between the homA or homB gene and clinical outcomes (Table 4).
hopQI and hopQII status
The hopQI gene was present in all strains examined, whereas only 6.9% of the strains carried the hopQII gene. The hopQII-positive strains were significantly more common in HLJ (13.6%) than in JX (2.9%) (P<0.001; Table 3). There was no hopQII-single positive strain in our study. Moreover, the frequency of the hopQI-single positive genotype was significantly higher in JX (97%) than in HLJ (86.4%, P<0.05; Table 3).
In the present study, no significant difference in hopQI nor hopQII distribution was observed between various diseases.
hrgA and hpyIIIR status
The dominant hpyIIIR-positive genotype was detected in 83.1% of the strains, which was significantly more prevalent than the hrgA-positive genotype (56.9%, P<0.001). We found that 41.9% of the strains were hpyIIIR-single positive genotypes, and 15.6% of the strains had hrgA in place of hpyIIIR. The mixed genotype for hrgA and hpyIIIR was common and accounted for 41.3% (Table 3).
The hpyIIIR gene was present in 88 (87.1%) strains from JX, which was more than that from HLJ (76.3%). In contrast, a significant difference was observed in the frequency of hrgA single-positive genotype between HLJ and JX (23.7% vs. 10.9%, P<0.001). The strains with mixed genotypes were significantly more common in JX (48.5%) than in HLJ (28.8%, P<0.001).
The hrgA-positive strains were more common in GC patients (71.1%) than in CG patients (50.7%), and the difference was statistically significant (P<0.05). In addition, 55.56% of PUD patients were infected with hrgA-positive strains; however, the difference was not statistically significant (P>0.05). The hpyIIIR-positive genotype was dominant, and the frequency was similar between various diseases. However, hypIIIR was detected in 92.6% of strains isolated from GC patients in JX, whereas only 63.6% of strains from HLJ were infected with hpyIIIR-positive strains. The difference was statistically significant (P<0.05). The hrgA single-positive genotype was present in 19.5% and 15.8% of strains from CG and GC patients, respectively, whereas only 8.9% of strains were detected in PUD patients. Moreover, the frequency of this genotype examined in GC strains was significantly different between HLJ and JX patients (36.4% and 7.4%, P<0.05). The frequency of the hpyIIIR single-positive genotype in strains isolated from CG patients (49.4%) was significantly higher than that in GC patients (28.9%, P<0.05; Table 4).
In our cohort, the mixed genotype was common and accounted for 55.3% and 31.2% of strains from GC and CG patients, respectively. The difference was statistically significant (P<0.05). Furthermore, the frequency of mixed genotypes in the CG strains (28.9%) isolated from JX was significantly lower than that in the PUD strains (52.8%, P<0.05) and GC strains (70.4%, P<0.001) (Table 4).
The relationship between different virulence factors
The association between different virulence factors was examined. homA and homB exhibited significant negative associations with each other (P<0.001), as did hrgA and hpyIIIR (P<0.001). A significant correlation between the presence of vacA m and hpyIIIR was observed (P<0.05).
When the virulence factors were combined, we found the hrgA/homB/vacA m2 and hrgA/homB/hpyIIIR genotypes were observed in 39.5% and 36.8% of the strains from GC patients, whereas only 3 strains were infected with the hrgA/homB/vacA m1 or hrgA/homB/jhp0563 genotype. The difference was statistically significant (P<0.001).
The relationship between gene status and clinical outcomes
Univariate analysis showed that only the hrgA gene was associated with GC. There was no significant association between the other genes and different diseases. The presence of the hrgA gene showed a significant association with GC and the risk of GC was 2.224-fold higher in patients infected by strains harboring the hrgA gene (95% CI: 1.014 to 4.882, P<0.05; Table 5).
Table 5
| Gene | GC | PUD | CG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |||
| vacA m1 | 0.706 | 0.313, 1.592 | 0.401 | 0.823 | 0.388, 1.747 | 0.613 | 1.491 | 0.764, 2.908 | 0.242 | ||
| m2 | 1.417 | 0.628, 3.198 | 0.401 | 1.215 | 0.572, 2.579 | 0.613 | 0.671 | 0.344, 1.309 | 0.242 | ||
| s1m1 | 0.732 | 0.324, 1.654 | 0.454 | 0.856 | 0.403, 1.820 | 0.688 | 1.409 | 0.720, 2.756 | 0.317 | ||
| s1m2 | 1.470 | 0.652, 3.314 | 0.353 | 1.263 | 0.596, 2.678 | 0.543 | 0.634 | 0.326, 1.235 | 0.181 | ||
| jhp0563 | 0.570 | 0.247, 1.314 | 0.185 | 1.388 | 0.674, 2.856 | 0.374 | 1.118 | 0.577, 2.167 | 0.743 | ||
| homA | 1.066 | 0.494, 2.301 | 0.871 | 0.762 | 0.359, 1.613 | 0.478 | 1.184 | 0.613, 2.288 | 0.617 | ||
| homB | 0.713 | 0.327, 1.556 | 0.396 | 1.352 | 0.615, 2.972 | 0.453 | 1.017 | 0.513, 2.018 | 0.962 | ||
| hopQII | 1.221 | 0.307, 4.855 | 0.777 | 0.955 | 0.242, 3.775 | 0.948 | 0.891 | 0.261, 3.048 | 0.855 | ||
| hrgA | 2.224 | 1.014, 4.882 | 0.044* | 0.928 | 0.463, 1.859 | 0.834 | 0.612 | 0.326, 1.149 | 0.127 | ||
| hpyIIIR | 1.109 | 0.412, 2.987 | 0.838 | 1.452 | 0.544, 3.873 | 0.456 | 0.699 | 0.304, 1.605 | 0.398 | ||
Univariate analysis shows the association between each gene status and the indicated diseases. *, indicate a significant difference. CG, chronic gastritis; PUD, peptic ulcer disease; GC, gastric carcinoma; OR, odds ratio; CI, confidence interval.
A multivariate analysis, including age, sex, and the status of different genes, was performed to determine the factors related to clinical outcomes. The results demonstrated that male patients were at significantly increased risk of GC compared to female patients [odds ratio (OR) =2.577, 95% CI: 1.028 to 6.865, P<0.05] and elderly patients were at a higher risk of GC development (OR =1.116, 95% CI: 1.075 to 1.168, P<0.05). Moreover, the hrgA genotype exhibited a positive correlation with GC and increased the risk of GC (OR =3.606, P<0.05; Table 6).
Table 6
| Disease | Gene | OR | 95% CI | P |
|---|---|---|---|---|
| GC | sex_Male | 2.577 | (1.028, 6.865) | 0.0489 |
| Age | 1.116 | (1.075, 1.168) | <0.0001 | |
| hrgA | 3.606 | (1.359, 10.071) | 0.0138 | |
| PUD | Age | 0.965 | (0.939, 0.991) | 0.0091 |
| CG | Age | 0.960 | (0.935, 0.984) | 0.0019 |
| sex_Male | 0.387 | (0.191, 0.765) | 0.0072 | |
| hrgA | 0.499 | (0.238, 1.024) | 0.0406 |
GC, gastric carcinoma; CG, chronic gastritis; PUD, peptic ulcer disease; OR, odds ratio; CI, confidence interval.
In contrast, the presence of hrgA exhibited a negative correlation with CG and represented a protective factor for CG (OR =0.499, P<0.05). In addition, sex and age also significantly decreased the risk of CG (OR =0.387, 95% CI: 0.191 to 0.765; and OR =0.960, 95% CI: 0.935 to 0.984, respectively; P<0.05). In patients with PUD, only age was observed to be associated with the disease, and the risk of PUD was 0.965-fold lower in elderly patients (OR =0.965, 95% CI: 0.939 to 0.991, P<0.05; Table 6).
Discussion
This current study demonstrated the distribution of distinct virulence genes in strains isolated from patients with various diseases and revealed a possible relationship with gastric diseases.
It is accepted that cagA is the most important virulence factor, and the prevalence of cagA ranges from 50% to 70% in European and American countries (32), whereas the prevalence is above 90% in some Asian countries (28,33). In addition, the prevalence of cagE ranges from 30 to 90% according to geographic regions (8,34,35). A recent study suggested that cagE combined with cagA can be used as marker of DU (8), however, their role in gastric pathogenesis remains unclear. In the present study, all strains tested were cagA and cagE positive, which may contribute to the higher incidence rate of GC in China compared to Western countries.
The vacA s1m1 genotype strains produce higher levels of toxins and are more highly associated with PUD and GC than the s2m2 genotype strains, which produce little or no toxins (11,36). After examining the vacA subtypes, we found that 98.7% of strains were of the s1 subtype, and strains containing m2 (68.1%) were more common, which is in contrast to previous studies (32,37), suggesting a difference between Chinese and foreign strains. The cagA and vacA s1 genotypes have been identified as markers of strains associated with DU or GC among Western populations (5). However, such a pattern is not observed in East Asia, where nearly all isolates have cagA and vacA s1 genotypes (28,38,39).
Almost all strains in our study cohort were positive for jhp0562, consistent with a study in Japan which reported the prevalence of jhp0562 was 100% and higher than that in the USA (12). Jhp0562 contributes to the synthesis of both type 1 and 2 Le antigens, which are expressed by East Asian strains, whereas the type 1 Le antigen is expressed by Western strains (40-42), and this may explain the geographical variations of jhp0562. In addition, two jhp0563 gene fragments of different lengths, which were observed in Western strains, were absent in our study cohort (profile 3 in Figure 2A). The length of jhp0563 varies due to the number of 21-nt tandem repeats, which is used as a gene switching mechanism (43,44). Studies have shown that jhp0562 is significantly associated with PUD and jhp0563 is associated with gastritis in some Western and American countries (12,13). Oleastro et al. reported a positive association between jhp0562 with cagA, likely leading to the strong link between jhp0562 with PUD (13). Moreover, jhp0562 and jhp0563 were found to describe the different incidences of GC (14). However, the present study did not reveal any association between the jhp0562 nor jhp0563 gene and clinical outcomes, consistent with studies in other Asian countries (12,14).
The homA and homB gene profiles vary greatly based on the geographic origin of the strains (45,46). Previous studies have shown that homB is found more frequently than homA in East Asian strains (19,30). In the present study, we observed that the positive rate of homB (71.3%) was significantly higher than that of homA (33.1%). The homB can increase secretion of proinflammatory cytokine interleukin (IL)-8 and increased inflammation is associated with more severe diseases, such as PUD and GC (47). The presence of homB is associated with PUD in Western strains, whereas homA is associated with non-ulcer diseases (48). However, we did not identify a relationship between homA nor homB and any disease type.
The homA and homB can be found interchangeably at two genomic loci, and various combinations of the genes can be found, including a single homA or homB gene, two copies of homA or homB, a single copy of each gene, or neither of these genes (18,49). Studies revealed that two copies-genotype of homB was strongly associated with PUD, therefore, the association between homB and disease is probably a result of the variations in genotype, geographic location, genomic loci and gene copy number in various strains (18,19,30). Further investigation is warranted to determine the copy number of these genes, as well as obtaining samples from other regions in China.
In our study, the presence of cagE, jhp0562, homB, and hopQI was prevalent, which probably attributed to the fact that the presence of these genes was common in cagA and vacA s1 strains (21,30,40,41,50). The differences in these gene profiles may contribute to the diversity of virulence between Chinese and Western strains and emphasize the characteristics of strains in higher-risk areas of GC, such as HLJ and JX. The strains harboring the cagA and vacA s1 genotypes could be considered highly virulent (49). Although we did not identify any relationship between the studied factors and different diseases in China, we also assumed that the presence of these genes could increase the virulence of strains in China. These factors can interact synergistically in some fashion and contribute to high levels of gastric mucosal inflammation, which is associated with the precancerous process (51,52).
In the present study, the frequency of hrgA (56.9%) in our strains was higher than that of other East Asian strains (e.g., 18% in Vietnam, 36% in Hong Kong, 31% in Korea), and similar to that of Western strains (e.g., 46% in Colombia, 55% in Canada) (27). It is worth noting that the mixed genotype for hpyIIIR and hrgA was observed in 41.25% of our strains, which was rare in other studies (25,27). Moreover, there was a significant difference in hrgA single-positive or mixed genotype between HLJ and JX strains, suggesting the obvious geographic diversity of this gene.
The frequency of hrgA-positive or mixed genotypes was significantly higher in GC strains than in CG strains, indicating that there was a strong correlation between these genotypes and GC. The significant difference was still apparent after multivariate analysis that adjusted for age, sex, and other factors. The multivariate analysis showed that the hrgA gene was independently associated with the development of GC and an increased risk of developing GC was associated with strains containing hrgA.
A previous study showed that there was no difference in the induction of IL-8 release nor apoptosis in gastric epithelial cells between hrgA and hpyIIIR strains (25). Therefore, whether the acquisition of hrgA is selected by GC pressure or is a causative factor was difficult to determine. We observed that the hpyIIIR single-positive genotype was more common in CG strains; moreover, the hrgA gene showed a negative association with CG. It is accepted that the development of GC begins with H. pylori-induced chronic superficial gastritis (53,54). Therefore, we speculated that the CG individuals with hrgA gene potentially develop GC due to interactions with host factors (e.g., single nucleotide polymorphisms of inflammatory cytokines) and environmental factors (38,55,56).
There were still some limitations in our research. First, the sample size of GC patients was small. Second, there were only two regions of China selected in our study. The distribution of virulence factors varied greatly in different regions of China. Although the two regions are representative of regional disparity, the 160 strains in our study couldn’t completely stand for the strains from other regions. Third, the function of HrgA protein and its role in GC progression is unclear. Further study is necessary to make adequate comparisons involving more GC patients from more regions of China to confirm the correlation between hrgA and GC, as well as to examine the function of HrgA. Once confirmed, hrgA may be used to identify individuals at higher GC risk in China.
Conclusions
In China, where there is a high prevalence of H. pylori infection and GC occurrence, the universal presence of studied virulence factors makes it impossible to examine disease-specific associations with any of these virulence factors. However, the distribution pattern of these factors suggested the characteristics of Chinese strains, which showed higher virulence and, ultimately, resulted in severe diseases. We found a strong association between hrgA status and progression to GC. We speculated that hrgA may serve as a promising candidate factor for severe disease development in China, and further study is warranted to confirm our speculation.
Acknowledgments
We would like to thank all the participants from the Department of Diagnosis for Communicable Diseases, the National Institute for Communicable Disease Control and Prevention, and the Chinese Center for Disease Control and Prevention.
Funding: This research was supported by the Research on Equipment for Rapid Diagnosis and Treatment of Marine “Superbug” Infection project (No. HJ20172B07021).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-23-1404/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-1404/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-1404/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-23-1404/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention (No. ICDC-2013001) and informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med 1991;325:1127-31. [Crossref] [PubMed]
- Šterbenc A, Jarc E, Poljak M, et al. Helicobacter pylori virulence genes. World J Gastroenterol 2019;25:4870-84. [Crossref] [PubMed]
- Santos A, Queiroz DM, Ménard A, et al. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J Clin Microbiol 2003;41:1651-5. [Crossref] [PubMed]
- Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci 2011;102:36-43. [Crossref] [PubMed]
- Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J 2016;39:14-23. [Crossref] [PubMed]
- Tomasini ML, Zanussi S, Sozzi M, et al. Heterogeneity of cag genotypes in Helicobacter pylori isolates from human biopsy specimens. J Clin Microbiol 2003;41:976-80. [Crossref] [PubMed]
- Khatoon J, Prasad KN, Prakash Rai R, et al. Association of heterogenicity of Helicobacter pylori cag pathogenicity island with peptic ulcer diseases and gastric cancer. Br J Biomed Sci 2017;74:121-6. [Crossref] [PubMed]
- El Khadir M, Boukhris SA, Zahir SO, et al. CagE, cagA and cagA 3’ region polymorphism of Helicobacter pylori and their association with the intra-gastric diseases in Moroccan population. Diagn Microbiol Infect Dis 2021;100:115372. [Crossref] [PubMed]
- Rhead JL, Letley DP, Mohammadi M, et al. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 2007;133:926-36. [Crossref] [PubMed]
- González CA, Figueiredo C, Lic CB, et al. Helicobacter pylori cagA and vacA genotypes as predictors of progression of gastric preneoplastic lesions: a long-term follow-up in a high-risk area in Spain. Am J Gastroenterol 2011;106:867-74. [Crossref] [PubMed]
- Sheikh AF, Yadyad MJ, Goodarzi H, et al. CagA and vacA allelic combination of Helicobacter pylori in gastroduodenal disorders. Microb Pathog 2018;122:144-50. [Crossref] [PubMed]
- Matsuda M, Shiota S, Matsunari O, et al. Prevalence of two homologous genes encoding glycosyltransferases of Helicobacter pylori in the United States and Japan. J Gastroenterol Hepatol 2011;26:1451-6. [PubMed]
- Oleastro M, Santos A, Cordeiro R, et al. Clinical relevance and diversity of two homologous genes encoding glycosyltransferases in Helicobacter pylori. J Clin Microbiol 2010;48:2885-91. [Crossref] [PubMed]
- Trang TT, Shiota S, Matsuda M, et al. The Prevalence of Helicobacter pylori Virulence Factors in Bhutan, Vietnam, and Myanmar Is Related to Gastric Cancer Incidence. Biomed Res Int 2015;2015:830813. [Crossref] [PubMed]
- Oleastro M, Monteiro L, Lehours P, et al. Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization. Infect Immun 2006;74:4064-74. [Crossref] [PubMed]
- Servetas SL, Kim A, Su H, et al. Comparative analysis of the Hom family of outer membrane proteins in isolates from two geographically distinct regions: The United States and South Korea. Helicobacter 2018;23:e12461. [Crossref] [PubMed]
- Alm RA, Bina J, Andrews BM, et al. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun 2000;68:4155-68. [Crossref] [PubMed]
- Oleastro M, Cordeiro R, Ferrand J, et al. Evaluation of the clinical significance of homB, a novel candidate marker of Helicobacter pylori strains associated with peptic ulcer disease. J Infect Dis 2008;198:1379-87. [Crossref] [PubMed]
- Jung SW, Sugimoto M, Graham DY, et al. homB status of Helicobacter pylori as a novel marker to distinguish gastric cancer from duodenal ulcer. J Clin Microbiol 2009;47:3241-5. [Crossref] [PubMed]
- Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol 2002;40:4504-11. [Crossref] [PubMed]
- Ohno T, Sugimoto M, Nagashima A, et al. Relationship between Helicobacter pylori hopQ genotype and clinical outcome in Asian and Western populations. J Gastroenterol Hepatol 2009;24:462-8. [Crossref] [PubMed]
- Cao P, Lee KJ, Blaser MJ, et al. Analysis of hopQ alleles in East Asian and Western strains of Helicobacter pylori. FEMS Microbiol Lett 2005;251:37-43. [Crossref] [PubMed]
- Chiarini A, Calà C, Bonura C, et al. Prevalence of virulence-associated genotypes of Helicobacter pylori and correlation with severity of gastric pathology in patients from western Sicily, Italy. Eur J Clin Microbiol Infect Dis 2009;28:437-46. [Crossref] [PubMed]
- Ando T, Aras RA, Kusugami K, et al. Evolutionary history of hrgA, which replaces the restriction gene hpyIIIR in the hpyIII locus of Helicobacter pylori. J Bacteriol 2003;185:295-301. [Crossref] [PubMed]
- Ando T, Wassenaar TM, Peek RM Jr, et al. A Helicobacter pylori restriction endonuclease-replacing gene, hrgA, is associated with gastric cancer in Asian strains. Cancer Res 2002;62:2385-9. [PubMed]
- G M. Association of Helicobacter pylori restriction endonuclease-replacing gene, hrgA with overt gastrointestinal diseases. Arq Gastroenterol 2008;45:225-9. [Crossref] [PubMed]
- Lu H, Graham DY, Yamaoka Y. The Helicobacter pylori restriction endonuclease-replacing gene, hrgA, and clinical outcome: comparison of East Asia and Western countries. Dig Dis Sci 2004;49:1551-5. [Crossref] [PubMed]
- Xue Z, Yang H, Su D, et al. Geographic distribution of the cagA, vacA, iceA, oipA and dupA genes of Helicobacter pylori strains isolated in China. Gut Pathog 2021;13:39. [Crossref] [PubMed]
- Proença Módena JL, Lopes Sales AI, Olszanski Acrani G, et al. Association between Helicobacter pylori genotypes and gastric disorders in relation to the cag pathogenicity island. Diagn Microbiol Infect Dis 2007;59:7-16. [Crossref] [PubMed]
- Oleastro M, Cordeiro R, Yamaoka Y, et al. Disease association with two Helicobacter pylori duplicate outer membrane protein genes, homB and homA. Gut Pathog 2009;1:12. [Crossref] [PubMed]
- Kong H, Lin LF, Porter N, et al. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res 2000;28:3216-23. [Crossref] [PubMed]
- van Doorn LJ, Figueiredo C, Sanna R, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 1998;115:58-66. [Crossref] [PubMed]
- Zhao Q, Song C, Wang K, et al. Prevalence of Helicobacter pylori babA, oipA, sabA, and homB genes in isolates from Chinese patients with different gastroduodenal diseases. Med Microbiol Immunol 2020;209:565-77. [Crossref] [PubMed]
- Asl SF, Pourvahedi M, Mojtahedi A, et al. Analysis of babA, cagE and cagA Genes in Helicobacter pylori from Upper Gastric Patients in the North of Iran. Infect Disord Drug Targets 2019;19:274-8. [Crossref] [PubMed]
- Markovska R, Boyanova L, Yordanov D, et al. Status of Helicobacter pylori cag pathogenicity island (cagPAI) integrity and significance of its individual genes. Infect Genet Evol 2018;59:167-71. [Crossref] [PubMed]
- Chauhan N, Tay ACY, Marshall BJ, et al. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: An overview. Helicobacter 2019;24:e12544. [Crossref] [PubMed]
- Yamaoka Y, Kodama T, Gutierrez O, et al. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol 1999;37:2274-9. [Crossref] [PubMed]
- Subsomwong P, Miftahussurur M, Uchida T, et al. Prevalence, risk factors, and virulence genes of Helicobacter pylori among dyspeptic patients in two different gastric cancer risk regions of Thailand. PLoS One 2017;12:e0187113. [Crossref] [PubMed]
- Hu Y, Wang Y, Mi M, et al. Correlation analysis of gastric mucosal lesions with Helicobacter pylori infection and its virulence genotype in Guiyang, Guizhou province, China. Ann Transl Med 2022;10:1320. [Crossref] [PubMed]
- Appelmelk BJ, Martino MC, Veenhof E, et al. Phase variation in H type I and Lewis a epitopes of Helicobacter pylori lipopolysaccharide. Infect Immun 2000;68:5928-32. [Crossref] [PubMed]
- Pohl MA, Romero-Gallo J, Guruge JL, et al. Host-dependent Lewis (Le) antigen expression in Helicobacter pylori cells recovered from Leb-transgenic mice. J Exp Med 2009;206:3061-72. [Crossref] [PubMed]
- Pohl MA, Zhang W, Shah SN, et al. Genotypic and phenotypic variation of Lewis antigen expression in geographically diverse Helicobacter pylori isolates. Helicobacter 2011;16:475-81. [Crossref] [PubMed]
- Salaün L, Linz B, Suerbaum S, et al. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology (Reading) 2004;150:817-30. [Crossref] [PubMed]
- Salaün L, Ayraud S, Saunders NJ. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiology (Reading) 2005;151:917-23. [Crossref] [PubMed]
- Oleastro M, Cordeiro R, Ménard A, et al. Allelic diversity and phylogeny of homB, a novel co-virulence marker of Helicobacter pylori. BMC Microbiol 2009;9:248. [Crossref] [PubMed]
- Fang M, Xue Z, He L, et al. Distribution characteristics of the sabA, hofC, homA, homB and frpB-4 genes of Helicobacter pylori in different regions of China. PLoS One 2022;17:e0268373. [Crossref] [PubMed]
- Atherton JC. The pathogenesis of Helicobacter pylori-induced gastroduodenal diseases. Annu Rev Pathol 2006;1:63-96. [Crossref] [PubMed]
- Kang J, Jones KR, Jang S, et al. The geographic origin of Helicobacter pylori influences the association of the homB gene with gastric cancer. J Clin Microbiol 2012;50:1082-5. [Crossref] [PubMed]
- Maeda S, Ogura K, Yoshida H, et al. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut 1998;42:338-43. [Crossref] [PubMed]
- Akar M, Kayman T, Abay S, et al. Clinical relevance of virulence genes in Helicobacter pylori isolates recovered from adult dyspeptic patients in Turkey. Indian J Med Microbiol 2022;40:258-62. [Crossref] [PubMed]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol 2010;7:629-41. [Crossref] [PubMed]
- Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res 1992;52:6735-40. [PubMed]
- Conteduca V, Sansonno D, Lauletta G, et al. H. pylori infection and gastric cancer: State of the art. Int J Oncol 2013;42:5-18. [Crossref] [PubMed]
- Matysiak-Budnik T, Mégraud F. Helicobacter pylori infection and gastric cancer. Eur J Cancer 2006;42:708-16. [Crossref] [PubMed]
- Figueiredo C, Machado JC, Pharoah P, et al. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J Natl Cancer Inst 2002;94:1680-7. [Crossref] [PubMed]
- Gong Y, Peng X, He L, et al. The distribution of jhp0940, jhp0945, jhp0947, jhp0949 and jhp0951 genes of Helicobacter pylori in China. BMC Gastroenterol 2015;15:115. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


