
Research reporting guidelines for cell lines: more than just a recommendation
Continuous growing cell lines are important experimental tools in biomedical research. However, despite their important role, evidence over the last decades has accumulated that many cell lines are frequently misidentified or cross-contaminated by other cells. The International Cell Line Authentication Committee (ICLAC) has recently launched version 12 of the register of misidentified cell lines (1). This register now lists 582 cross-contaminated or misidentified cell lines. Of these cell lines, 21 were initially thought to be of hepatic origin, but later shown to be contaminated by HeLa (human cervical adenocarcinoma cell line), HCT8 (human colon carcinoma cell line), HepG2 (human hepatoblastoma cell line), RAW 264.7 (mouse leukemia cell line), or with cells of unknown origin. Although the problems and effects of the use of falsified cells in biomedical research are well-known, several of these cell lines are still frequently used in hepatology research (Table 1).
Table 1
| Cell line/(Cellosarus ID) | ICLAC registration ID | Claimed species | Claimed cell type | Contaminating cell line | Actual species | Actual cell line | References | Search term/number of publications‡ |
|---|---|---|---|---|---|---|---|---|
| BEL-7402 (CVCL_5492) | ICLAC-00549 | Human | Liver, hepatocellular carcinoma | HeLa/HCT 8 | Human | Cervical adenocarcinoma/colon carcinoma | (2-5) | “BEL-7402”/1371 |
| BEL-7404 (CVCL_6568) | ICLAC-00550 | Human | Liver, hepatocellular carcinoma | HeLa | Human | Cervical adenocarcinoma | (3,5) | “BEL-7404”/261 |
| Chang liver (CVCL_0238) | ICLAC-00002 | Human | Liver, normal hepatic cells | HeLa | Human | Cervical adenocarcinoma | (6-8) | “Chang liver” or “Changliver”/702 |
| D-11 (R1 derivative) (CVCL_2012) | ICLAC-00582 | Rainbow trout | Liver, normal hepatic cells | Unknown | Chinook salmon | Unknown | (9) | “D-11 cell”/1 |
| GREF-X (CVCL_7667) | ICLAC-00123 | Human | Liver, hepatic myofibroblast | Unknown | Rat | Unknown | (10) | “GREF-X”/1 |
| H7D7A (CVCL_1T06) | ICLAC-00203 | Human | Liver, normal cells (SV40-transformed) | HepG2 | Human | Liver, hepatoblastoma | (11) | “H7D7A”/0 |
| H7D7B (CVCL_1T07) | ICLAC-00204 | Human | Liver, normal cells (SV40-transformed) | HepG2 | Human | Liver, hepatoblastoma | (11) | “H7D7B”/0 |
| H7D7BD5 (H7D7B derivative) (CVCL_1T10) | ICLAC-00560 | Human | Liver, normal cells (SV40-transformed) | HepG2 | Human | Liver, hepatoblastoma | (11) | “H7D7BD5”/0 |
| H7D7C (CVCL_1T08) | ICLAC-00205 | Human | Liver, normal cells (SV40-transformed) | HepG2 | Human | Liver, hepatoblastoma | (11) | “H7D7C”/0 |
| H7D7D (CVCL_1T09) | ICLAC-00206 | Human | Liver, normal cells (SV40-transformed) | HepG2 | Human | Liver, hepatoblastoma | (11) | “H7D7D”/0 |
| Hepa-T1 (CVCL_4226) | ICLAC-00567 | Nile tilapia | Liver, normal hepatic cells | Unknown, possibly Hepa-E1 | Japanese eel | Unknown | (12) | “Hepa-T1”/8 |
| HuL-1 (CVCL_8357) | ICLAC-00318 | Human | Liver, hepatocellular carcinoma | HeLa | Human | Cervical adenocarcinoma | (13) | “HuL-1 and liver”/5 |
| ImKC (CVCL_HF55) | ICLAC-00620 | Mouse [H-2K(b)-tsA58 transgenic line] | Liver, normal Kupffer cells | RAW 264.7 | Mouse | Macrophage, transformed | (1) | “ImKC” and “Kupffer or macrophage”/2 |
| L-02 (CVCL_6926) | ICLAC-00575 | Human | Liver, normal hepatic cells | HeLa | Human | Cervical adenocarcinoma | (3) | “LO2 cell or L-O2 cell” and “liver”/562 |
| QGY-7701 (CVCL_6859) | ICLAC-00551 | Human | Liver, hepatocellular carcinoma | HeLa | Human | Cervical adenocarcinoma | (4,5) | “QGY-7701”/49 |
| QGY-7703 (CVCL_6715) | ICLAC-00552 | Human | Liver, hepatocellular carcinoma | HeLa | Human | Cervical adenocarcinoma | (4,5) | “QGY-7703”/125 |
| QSG-7701 (CVCL_6944) | ICLAC-00553 | Human | Liver, normal hepatic cells | HeLa | Human | Cervical adenocarcinoma | (2,5) | “QSG-7701”/59 |
| R1 (CVCL_4607) | ICLAC-00581 | Rainbow trout | Liver, normal hepatic cells | Unknown | Chinook salmon | Unknown | (14) | “R1 cell” and “liver”/1 |
| RBHF-1 (CVCL_Y465) | ICLAC-00155 | Human | Liver, hepatoma | Unknown | Non-human | Unknown | (10) | “RBHF-1”/1 |
| SMMC-7721 (CVCL_0534) | ICLAC-00554 | Human | Liver, hepatocellular carcinoma | HeLa | Human | Cervical adenocarcinoma | (2,5) | “SMMC-7721”/2332 |
| WRL 68 (CVCL_0581) | ICLAC-00351 | Human | Liver, embryonic cells | HeLa | Human | Cervical adenocarcinoma | (15) | “WRL 68 or WRL68”/248 |
†, information of the first 8 columns of this table was taken in modified from the latest ICLAC register (version 12) that were released on 16 January 2023 (1); ‡, to estimate the usage of the different cell lines a search was conducted on 6th March, 2023 using the given keyword terms in the PubMed database (16). ICLAC, International Cell Line Authentication Committee.
For example, the cell line SMMC-7721 (CVCL_0534), also known as H-7721, was originally established from a Chinese male patient with hepatocellular carcinoma in 1977 (17), but 38 years later it was suggested to be contaminated and taken over either by HeLa and/or by a second cross-contaminating intruder introduced during prolonged culturing (2). Although unmasked nearly two decades ago as a cell line that was not derived from the liver, this cell line is still incomprehensibly used at high frequency in hepatology research (Figure 1).
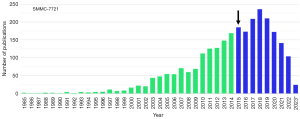
In most of these studies this cell line is either called a “human liver cancer cell line”, “human hepatoma cell line”, “carcinoma cell line”, “human hepatocellular carcinoma cell line”, “liver cancer cell”, or as “hepatocellular carcinoma (HCC) line”. Unfortunately, the cells are also used in a preclinical “SMMC-7721 xenograft nude mouse model” established to assess the efficacy of novel drugs that might be beneficial for liver cancer patients. This cell-line-derived xenograft (CDX) model is advertised as a model that “offers key decision-making information” (18) or “is essential for biomedical research related to human liver cancer” (19).
Similarly, the Chang liver cell (CVCL_0238) is a widely used misidentified cell line. Originally established from a normal human liver biopsy, it was introduced as a human “normal hepatocyte model” (20). Twenty-two years later it was realized that this cell line is cross-contaminated with HeLa cells (7). Nevertheless, some researchers still uses this cell line as a “healthy normal liver cell” model. Another prominent example of a misidentified liver cell line is L-O2 (CVCL_6926) that was originally established in 1980 at the Chinese Academy of Sciences (Shanghai) by immortalization of a primary normal human hepatocyte with the human telomerase reverse transcriptase (hTERT) (21). In line with this assumption, the cells were shown to maintain biological features and the ultrastructure of normal adult hepatocytes (21). Moreover, it was demonstrated that this non-tumor hepatic cell line exhibited good liver function properties, expressed albumin, and further improved liver function in rats subjected to hepatotectomy (22). However, it is not known if this cell line used in this study was misidentified or cross contaminated. Later, however it was proven by genetic profiling that this cell line (which is also termed “Liver-02”, “L-02”, “LO2”, “human liver-7702”, or “HL-7702”) is a derivative of HeLa (2). It is therefore extremely surprising that this cell line is still used as a normal liver cell line in many studies. A recent search at the PubMed database using the search terms “LO2 cell or L-O2 cell” and “liver” conducted on 8 March 2023 resulted in 562 hits and revealed that this cell line is being used more and more from year to year (e.g., 47 studies in 2017, 73 studies in 2019, 83 studies in 2021). Nevertheless, clearly stated, the 21 misidentified liver cell lines listed by the ICLAC must be classified as unusable for hepatology research.
False cell lines are a widespread problem in biomedical research. The general prevalence of misidentified cell lines and their impact on reproducibility has recently been discussed (23). The authors mentioned that at least 5% of cell lines in manuscripts considered for peer-review that were submitted to a highly reputable cancer journal (i.e., International Journal of Cancer) were misidentified. Interestingly, the majority of papers that were rejected by the journal for serious cell line-related problems were published in other journals (23), demonstrating that many authors just ignore that they publish potentially artifactual or faulty research results.
Unfortunately, the identification of a misidentified cell line is complicated because cell lines often have several synonyms, just like in the case of L-O2 or SMMC-7721. In addition, workflows for quality control and cell line authentication testing should be established in laboratories working with established cell lines (24). In particular, cell line identity testing using short tandem repeat (STR) profiling is an effective means to prevent cell line misidentification and to identify cross-contamination at early stage (24). Guidelines and guidance for proper documentation of experiments that are conducted with continuous cell lines are published elsewhere (25). Necessary information to be reported includes the proper cell line name, supply source including order number (as listed in cell bank catalogues), a brief description of the cell line (species, age and sex, strain, tissue source, passage number), morphological and growth characteristics, culture conditions (media and supplements), information about quality control and cell authentication, hazard classification, and a citation to the paper in which the establishment of the respective cell line under investigation was described (25). Unfortunately, the awareness and knowledge about the problems associated with misidentified cell lines is rather low (23). Nevertheless, strict research reporting guidelines for cell lines, to which serious scientists adhere to, should be mandatory to prevent publication of faulty research data.
There is only hope that better training in good cell culture practice (GCCP) and the implementation of more strict guidelines in authentication policies by funding agencies, research institutions, and journals will reduce the number of papers reporting results with misidentified or cross-contaminated cell lines (25). Experts have provided several landmark recommendations for characterizing cell lines in biomedical research that are extremely helpful for those working with established cell lines (26-29).
Actually, the peer review process from many journals has mostly failed to detect and monitor the use of falsely designated cell lines such as SMMC-7721, Chang liver cells, LO-2, and many others. In addition, more critical reviewers are urgently needed that will control the minimal reporting requirements in studies using continuous cell lines, thereby sustainably correcting and preventing the enormous problems that are associated with false, non-reproducible research results. In this context, the constantly updated lists of the ICLAC and the Cellosaurus database that currently contains (release 44 of December 12, 2022) extensive information on 144,568 cell lines from 805 species are particularly helpful for reviewers to quickly identify unusable cell lines (1,30-32).
It would possible to prevent the publication of unreliable, potentially meaningless results when funding agencies and research journals would be more strict in requesting precise sources of cells and information about strategies and frequencies of authentication tests during work with continuously growing cell lines. It would be helpful if journals and agencies would provide an example of the template used by peer reviewers of National Institutes of Health (NIH) (or other non-US funding agency) grant applications to illustrate the current requirements for providing information/data on cell line authentication. Most recently, the ICLAC has published a Cell Line Checklist for Manuscript and Grant Applications in which standards for reporting were summarized (33).
Minimal reporting requirements in this Checklist for cell lines are the Cellosaurus (CVCL)/resource identification (RRID) identification number, strategy used for authentication, results of testing for mycoplasma, source of cell line, and information about the outcome of replicate experiments. Consequently, researchers should establish a work strategy in which registers of known misidentified cell lines are regularly screened, authentication tests established for new cell lines established, and respective results are comprehensibly reported in publications and grant applications. The Editors in Chief of scientific journals and their associated staff (Associate Editors, Editorial Board members) should be responsible for providing and verifying the compliance of clear guidelines to each peer reviewer requiring the inclusion of data demonstrating authentication results. Likewise, the relevance of cell line authentication for grant application is also emphasized by the NIH in Notice number NOT-OD-08-017 (34), NOT-OD-15-103 (35), and the Infographics on grant guidelines (36). These guidelines attempt to promote rigorous and transparent research in all research areas that address reproducibility, rigor, and transparency.
Regrettably, most reagents, tools and protocols for cell authentication are currently mostly limited to human cell lines. In this regard the Assay Guidance Manual has recently published a chapter on authentication of human and mouse cell lines (26). This chapter provides important guidelines for researchers who will need to interpret STR genotyping data generated in their laboratory or received from a core facility or commercial testing laboratory for the authentication of mouse and human cell lines. It further provides information about the workflow, demanding experimental steps, and troubleshooting guide for all critical steps in STR profiling.
So what argues against accepting and following these guidelines to foster reproducibility in biomedical research?
Acknowledgments
Funding: The laboratory of RW is supported by
Footnote
Provenance and Peer Review: This article was a standard submission to the journal. The article has undergone external peer review.
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-23-1208/prf
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-23-1208/coif). R.W. serves as an unpaid editorial board member of Annals of Translational Medicine from August 2022 to July 2024. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- ICLAC Register of Misidentified Cell Lines. Available online: https://iclac.org/ (last accessed 9 March 2023).
- Ye F, Chen C, Qin J, et al. Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China. FASEB J 2015;29:4268-72. [Crossref] [PubMed]
- Huang Y, Liu Y, Zheng C, et al. Investigation of Cross-Contamination and Misidentification of 278 Widely Used Tumor Cell Lines. PLoS One 2017;12:e0170384. [Crossref] [PubMed]
- Bian X, Yang Z, Feng H, et al. A Combination of Species Identification and STR Profiling Identifies Cross-contaminated Cells from 482 Human Tumor Cell Lines. Sci Rep 2017;7:9774. [Crossref] [PubMed]
- Rebouissou S, Zucman-Rossi J, Moreau R, et al. Note of caution: Contaminations of hepatocellular cell lines. J Hepatol 2017;67:896-7. [Crossref] [PubMed]
- Gartler SM. Genetic markers as tracers in cell culture. Natl Cancer Inst Monogr 1967;26:167-95.
- Lavappa KS, Macy ML, Shannon JE. Examination of ATCC stocks for HeLa marker chromosomes in human cell lines. Nature 1976;259:211-3. [Crossref] [PubMed]
- Nelson-Rees WA, Daniels DW, Flandermeyer RR. Cross-contamination of cells in culture. Science 1981;212:446-52. [Crossref] [PubMed]
- DSMZ – German Collection of Microorganisms and Cell Cultures GmbH. D-11 – Discontinued. Available online: https://www.dsmz.de/collection/catalogue/details/culture/ACC-77 (last accessed 9 March 2023).
- MacLeod RA, Dirks WG, Matsuo Y, et al. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer 1999;83:555-63. [Crossref] [PubMed]
- van Pelt JF, Decorte R, Yap PS, et al. Identification of HepG2 variant cell lines by short tandem repeat (STR) analysis. Mol Cell Biochem 2003;243:49-54. [Crossref] [PubMed]
- Tanaka R, Hatate H, Ito M, et al. Correction to: Elevation of lipid peroxide level and production of hydroxy lipids in cultured Hepa-T1 cells by oxidative stressors. Fish Sci 2018;84:733-4.
- Cellosaurus HuL-1 (CVCL_8357). Available online: https://www.cellosaurus.org/CVCL_8357 (last accessed 9 March 2023).
- DSMZ – German Collection of Microorganisms and Cell Cultures GmbH. R1. Available online: https://www.dsmz.de/collection/catalogue/details/culture/ACC-56 (last accessed 9 March 2023).
- European Collection of Authenticated Cell Cultures (ECACC). Available online: https://www.culturecollections.org.uk/products/celllines/generalcell/detail.jsp?refId=89121403&collection=ecacc_gc (last accessed 9 March 2023).
- PubMed. National Library of Medicine. Available online: https://pubmed.ncbi.nlm.nih.gov/ (last accessed 9 March 2023).
- Dong RC. Establishment of a human hepatocarcinoma cell line SMMC-7721 and initial observations on its biologic characteristics. In: Tang ZY, Wu MC, Xia SS (editors) Primary liver cancer, editors. Berlin: Springer; 1989. pp. 145–153. ISBN-13: 9780387502281.
- Creative Bioarray. Available online: https://dda.creative-bioarray.com/cell-based-xenograft-models.html (last accessed 9 March 2023).
- Altogen labs. SMMC-7721 xenograft model. Available online: https://altogenlabs.com/SMMC-7721_Xenograft_Service.pptx (last accessed 9 March 2023).
- CHANG RS. Continuous subcultivation of epithelial-like cells from normal human tissues. Proc Soc Exp Biol Med 1954;87:440-3. [Crossref] [PubMed]
- Ye XZ, Zhu DH, Shen DW. Ultrastructures of LO2 normal adult hepatocytes over successive in vitro cultures. Acta Biol Exp Sin 1980;13:3612-41. (Shi Yan Sheng Wu Xue Bao).
- Hu X, Yang T, Li C, et al. Human fetal hepatocyte line, L-02, exhibits good liver function in vitro and in an acute liver failure model. Transplant Proc 2013;45:695-700. [Crossref] [PubMed]
- Souren NY, Fusenig NE, Heck S, et al. Cell line authentication: a necessity for reproducible biomedical research. EMBO J 2022;41:e111307. [Crossref] [PubMed]
- Weiskirchen S, Schröder SK, Weiskirchen R. A beginner’s guide to cell culture: Practical advice for preventing needless problems. Cells 2023;12:682. [Crossref] [PubMed]
- Pamies D, Leist M, Coecke S, et al. Guidance document on Good Cell and Tissue Culture Practice 2.0 (GCCP 2.0). ALTEX 2022;39:30-70. [Crossref] [PubMed]
- Almeida JL, Korch CT. Authentication of Human and Mouse Cell Lines by Short Tandem Repeat (STR) DNA Genotype Analysis. In: Markossian S, Grossman A, Brimacombe K, et al., eds. Assay Guidance Manual. Bethesda (MD): Eli Lilly & Company and the National Center for Advancing Translational Sciences; January 17, 2023.
- Geraghty RJ, Capes-Davis A, Davis JM, et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer 2014;111:1021-46. [Crossref] [PubMed]
- Korch CT, Hall EM, Dirks WG, et al. 2022. Human Cell Line Authentication. Standardization of Short Tandem Repeat (STR) Profiling. ASN-0002 Revised 2022, November 2022 ed. American National Standards Institute (ANSI) - American Type Culture Collection (ATCC) Standards Development Organization, Manassas, Virginia, United States. Available online: https://webstore.ansi.org/standards/atcc/ansiatccasn00022022
- Freshney's Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications, 8th edition by R. Ian Freshney and Amanda Capes-Davis. 2021. Published by Wiley/Blackwell. ISBN: 978-1-119-51304-9.
- Bairoch A. The Cellosaurus, a Cell-Line Knowledge Resource. J Biomol Tech 2018;29:25-38. [Crossref] [PubMed]
- Cellosaurus – a knowledge resource on cell lines. Available online: https://www.cellosaurus.org/ (last accessed 9 March 2023).
- Cellosaurus newsletter 10 of January 2023. Available online: https://www.cellosaurus.org/news_archive/cellosaurus_news_10.pdf (last accessed 9 March 2023).
- International Cell Line Authentication Committee (ICLAC). Cell Line Checklist for Manuscripts and Grant Applications. Available online: https://iclac.org/wp-content/uploads/ICLAC_Cell-Line-Checklist_03-Mar-2023.pdf (last accessed 19 May 2023).
- NIH. NOT-OD-08-017. Notice Regarding Authentication of Cultured Cell Lines. Available online: https://grants.nih.gov/grants/guide/notice-files/not-od-08-017.html (last accessed 19 May 2023).
- NIH. NOT-OD-15-103. Enhancing Reproducibility through Rigor and Transparency. Available online: https://grants.nih.gov/grants/guide/notice-files/not-od-15-103.html (last accessed 19 May 2023).
- NIH. Grants & Funding. Central Resource for Grants and Funding Information. Available online: https://grants.nih.gov/policy/reproducibility/guidance.htm (last accessed 19 May 2023).


