
Mechanism and ingredients prediction of Radix Salviae-Angelicae Sinensis Radix-Lycii Fructus-Rehmanniae Radix Praeparata-Ginkgo Folium for retinitis pigmentosa therapy using network pharmacology and molecular docking analysis
Highlight box
Key findings
• We thoroughly investigated the mechanism of Radix Salviae-Angelicae Sinensis Radix-Lycii Fructus-Rehmanniae Radix Praeparata-Ginkgo Folium (RALRG) for retinitis pigmentosa (RP). Luteolin, quercetin, and kaempferol may be promising ingredients of RALRG for RP treatment.
What is known and what is new?
• The treatment of RP has become more demanding recently and RALRG are commonly used herbs in China. RALRG exert some protection on retinal cells independently.
• The impact of RALRG on RP was researched.
What is the implication, and what should change now?
• This study provides a new perspective for comprehensive treatment in RP by paying attention to Chinese herbs and provides a theoretical reference for the next research of applying RALRG in RP.
• Luteolin, quercetin, and kaempferol may be applied to further research the protective effect in vivo and in vitro.
Introduction
Retinitis pigmentosa (RP), the most common form of retinal degeneration, is characterized by the progressive loss of rods and cones, affecting approximately 1 in 4,000 individuals (1,2). Mutations in more than 80 genes have been implicated in non-syndromic RP, and every year new genes are added to this list (3). The heterogeneity of genetic mutations makes treatment of RP complex and difficult (2,4,5). Pharmacological treatments may delay the progression of photoreceptor and retinal pigment epithelium (RPE) cell loss with the benefit of few adverse effects (2). Therefore, the discovery of effective drugs targeting the common pathological process of RP is crucial.
RP belongs to the ‘vision of the high-altitude wind sparrow category’ in traditional Chinese medicine (TCM) (6). TCM has shown many advantages in improving visual acuity and the visual field (6). Radix Salviae (Danshen)-Angelicae Sinensis Radix (Danggui)-Lycii Fructus (Gouqizi)-Rehmanniae Radix Praeparata (Shudihuang)-Ginkgo Folium (Yinxinye) (together known as RALRG) are five herbs that have been long been commonly used in the treatment of the vision of high-altitude wind sparrow in China (6,7). Recent studies have tested some of the aforementioned herbs on animals to explore the mechanism. For example, Lycii Fructus and Radix Salviae extract was shown to alleviate RP through the Nrf2/HO-1 signaling pathway in rd10 mice (7), and Ginkgo Folium was found to delay light-induced photoreceptor degeneration through antioxidant and antiapoptotic properties (8). However, due to the multiple components and complex effects of Chinese herbs, the involved active components and their mechanism of action are still unknown. Network pharmacology is a new method that combines computer science and medicine to determine the therapeutic mechanism of drugs by constructing a visualized “multi-gene, multi-target, multi-pathway” interaction network, which is very suitable for the research of multi-component Chinese herbs (9,10). Molecular docking is an established in silico, structure-based method that refers to combining small molecules with macromolecules through binding sites, which is widely used in drug discovery (11).
We speculated that the components of RALRG have a specific protective effect on RP. Thus, the targets of active ingredients of RALRG were analyzed and summarized. Then, through network pharmacology, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, the mechanism of the active components of RALRG in treating RP was identified. The promising active ingredients in RALRG were analyzed using molecular docking technology. This study aimed to provide a theoretical basis for screening relevant molecules in treating RP and understanding the molecular mechanism. The workflow is shown in Figure 1. We present this article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3557/rc).
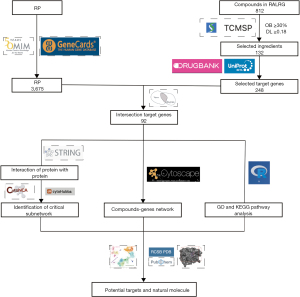
Methods
Establishing RALRG active ingredient database
The ingredients of RALRG were searched and collected from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP; https://old.tcmsp-e.com/index.php) (12). The TCMSP database contains all 499 Chinese medicines registered by the Chinese Pharmacopoeia, including 29,384 components, 3,311 targets, and 837 related diseases. It also provides pharmacokinetic properties such as oral bioavailability (OB), drug-likeness (DL), intestinal epithelial permeability, blood-brain barrier penetrability, and water solubility. Active ingredient molecular structures and detailed pharmacological profiles were retrieved from the PubChem database (http://pubchem.ncbi.nlm.nih.gov) (13).
Clustering RP- and RALRG-related target genes
RP-related target genes were downloaded from GeneCards (https://www.genecards.org/) (14) and Online Mendelian Inheritance in Man (OMIM; https://omim.org/search/advanced/geneMap) (15). To reduce the false positives, only the mutated genes identified in RP patients and the genes regulated in RP patients, which expressed in the retina [found in the human protein atlas (HPA) database; https://www.proteinatlas.org] and were directly associated with RP were included. The RALRG-related target genes were clustered depending on chemical similarities and pharmacophore models via the TCMSP database. These RALRG-related target names were calibrated to standardized names using DrugBank (https://go.drugbank.com/) database (16) and the UniProt database (https://www.uniprot.org) (17). Potential target genes of RALRG for RP were acquired through the Venn diagram intersection (https://bioinformatics.psb.ugent.be/webtools/Venn/) and their experimental support data were acquired from the Comparative Toxicogenomics Database (CTD; https://ctdbase.org/).
Integration of protein-protein interaction (PPI) network
A PPI network map was constructed for the co-expression, fusion, neighborhood, and co-localization of potential target genes with predicted gene interactions via the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) with “Homo sapiens” selected (18). The top 10 genes were screened in the PPI network using the Matthews correlation coefficient (MCC) algorithm based on the CytoHubba plugin without checking the first-stage nodes. The CytoNCA plugin in Cytoscape was also used to screen key sub-networks (19). Genes were filtered according to the primary score file calculated by CytoNCA that each score of betweenness centrality (BC), closeness centrality, degree centrality (DC), eigenvector centrality (EC), local average connectivity (LAC), and network centrality (NC) was higher than the median value. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Functional enrichment analysis
The R package “org.Hs.eg.db” was used to transform the gene symbol to Entrez ID. GO biological functions and KEGG pathway enrichment were analyzed by the R packages “DOSE”, “clusterProfiler”, and “pathview”, and visualized by the “enrichplot” package. R software (R version 4.1.3; https://cran.r-project.org/) was used for all statistical analyses. The significant enrichment threshold was set as P adjust <0.05.
Single-cell analysis of key target genes and binding capacity between active ingredients and key target genes
The single-cell profiles were downloaded from the HPA database. Cell clustering was performed to classify into five cell types and the expression of key target genes was counted. The three-dimensional (3D) structure of the proteins was downloaded from the Protein Data Bank (PDB) database (https://www.pdbus.org) (20), and the existing water molecules and nucleic acid strand small molecules were removed. The docking of macromolecules and ligands was analyzed using CB-Dock online molecular docking (http://cao.labshare.cn/cb-dock) based on the curvature-based cavity detection approach (CurPocket) and the AutoDock Vina to facilitate the docking procedure and improve the accuracy (21). After determining the docking pocket coordinates, molecular docking and conformation scoring were performed. The lower the score is, the more stable the binding of the ligand to the macromolecules is, which can preliminarily evaluate the binding activity of the ingredients to the target.
Statistical analysis
All statistical tests were analyzed in the R software (R version 4.1.3) and the Cytoscape (version 3.9.1; https://cytoscape.org/). The specific calculation was described in each step. Statistical significance was defined at two-tailed P<0.05.
Results
RALRG active ingredients database establishment
A total of 812 ingredients were searched and collected from RALRG. Based on the screening conditions of OB ≥30% and DL ≥0.18 (12), 132 ingredients were selected from the RALRG ingredients for database establishment after deleting duplication (Table 1). The highest OB value of these compounds was przewalskin b, which was 110.32, and the largest DL value was (24R)-4alpha-Methyl-24-ethylcholesta-7,25-dien-3beta-ylacetate, which was 0.84.
Table 1
| No. | MOL ID | Molecule name | OB (%) | MW | DL |
|---|---|---|---|---|---|
| 1 | MOL007156 | Przewalskin b | 110.32 | 330.46 | 0.44 |
| 2 | MOL009618 | (2R)-3-(3,4-dihydroxyphenyl)-2-[(Z)-3-(3,4-dihydroxyphenyl)acryloyl]oxy-propionic acid | 109.38 | 360.34 | 0.35 |
| 3 | MOL009678 | Physalin A | 91.71 | 526.58 | 0.27 |
| 4 | MOL009681 | (Z)-3-[2-[(E)-2-(3,4-dihydroxyphenyl)vinyl]-3,4-dihydroxy-phenyl]acrylic acid | 88.54 | 314.31 | 0.26 |
| 5 | MOL009617 | Bilobalide | 84.42 | 326.33 | 0.36 |
| 6 | MOL007145 | (6S)-6-hydroxy-1-methyl-6-methylol-8,9-dihydro-7H-naphtho[8,7-g]benzofuran-10,11-quinone | 75.39 | 312.34 | 0.46 |
| 7 | MOL000449 | Formyltanshinone | 73.44 | 290.28 | 0.42 |
| 8 | MOL000358 | Miltionone II | 71.03 | 312.39 | 0.44 |
| 9 | MOL009642 | Epidanshenspiroketallactone | 68.27 | 284.38 | 0.31 |
| 10 | MOL007154 | (6S)-6-(hydroxymethyl)-1,6-dimethyl-8,9-dihydro-7H-naphtho[8,7-g]benzofuran-10,11-dione | 65.26 | 310.37 | 0.45 |
| 11 | MOL009639 | Prolithospermic acid | 64.37 | 314.31 | 0.31 |
| 12 | MOL005438 | 2-(4-hydroxy-3-methoxyphenyl)-5-(3-hydroxypropyl)-7-methoxy-3-benzofurancarboxaldehyde | 62.78 | 356.40 | 0.40 |
| 13 | MOL001323 | Przewaquinone B | 62.24 | 292.30 | 0.41 |
| 14 | MOL000098 | Digallate | 61.85 | 322.24 | 0.26 |
| 15 | MOL007130 | Flavoxanthin | 60.41 | 584.96 | 0.56 |
| 16 | MOL007127 | Danshenol B | 57.95 | 354.48 | 0.56 |
| 17 | MOL007141 | Danshenol A | 56.97 | 336.41 | 0.52 |
| 18 | MOL001771 | Sesamin | 56.55 | 354.38 | 0.83 |
| 19 | MOL001494 | Przewaquinone c | 55.74 | 296.34 | 0.40 |
| 20 | MOL007449 | Isocryptotanshi-none | 54.98 | 296.39 | 0.39 |
| 21 | MOL011586 | (+)-catechin | 54.83 | 290.29 | 0.24 |
| 22 | MOL009631 | Neocryptotanshinone | 52.49 | 314.41 | 0.32 |
| 23 | MOL007152 | Tanshinaldehyde | 52.47 | 308.35 | 0.45 |
| 24 | MOL006209 | Cryptotanshinone | 52.34 | 296.39 | 0.40 |
| 25 | MOL011589 | Glycitein | 50.48 | 284.28 | 0.24 |
| 26 | MOL009640 | Danshenspiroketallactone | 50.43 | 282.36 | 0.31 |
| 27 | MOL005406 | Isotanshinone II | 49.92 | 294.37 | 0.40 |
| 28 | MOL001979 | Tanshinone iia | 49.89 | 294.37 | 0.40 |
| 29 | MOL009634 | Miltionone I | 49.68 | 312.39 | 0.32 |
| 30 | MOL011587 | (−)-catechin | 49.68 | 290.29 | 0.24 |
| 31 | MOL009641 | Isorhamnetin | 49.60 | 316.28 | 0.31 |
| 32 | MOL009653 | Deoxyneocryptotanshinone | 49.40 | 298.41 | 0.29 |
| 33 | MOL007061 | Ginkgolide M | 49.09 | 424.44 | 0.75 |
| 34 | MOL007101 | Ginkgolide C | 48.33 | 440.44 | 0.73 |
| 35 | MOL009635 | (E)-3-[2-(3,4-dihydroxyphenyl)-7-hydroxy-benzofuran-4-yl]acrylic acid | 48.24 | 312.29 | 0.31 |
| 36 | MOL007045 | Cyanin | 47.42 | 411.66 | 0.76 |
| 37 | MOL007125 | Cryptoxanthin monoepoxide | 46.95 | 568.96 | 0.56 |
| 38 | MOL007119 | 6-o-syringyl-8-o-acetyl shanzhiside methyl ester | 46.69 | 628.64 | 0.71 |
| 39 | MOL009664 | Quercetin | 46.43 | 302.25 | 0.28 |
| 40 | MOL001601 | (24R)-4alpha-Methyl-24-ethylcholesta-7,25-dien-3beta-ylacetate | 46.36 | 482.87 | 0.84 |
| 41 | MOL005573 | Ethyl linolenate | 46.10 | 306.54 | 0.20 |
| 42 | MOL007179 | Linolenic acid ethyl ester | 46.10 | 306.54 | 0.20 |
| 43 | MOL000096 | Atropine | 45.97 | 289.41 | 0.19 |
| 44 | MOL009646 | Tanshinone VI | 45.64 | 296.34 | 0.30 |
| 45 | MOL007123 | Salvianolic acid g | 45.56 | 340.30 | 0.61 |
| 46 | MOL000354 | Isoimperatorin | 45.46 | 270.30 | 0.23 |
| 47 | MOL000492 | Manool | 45.04 | 304.57 | 0.20 |
| 48 | MOL009612 | DihydrotanshinoneⅠ | 45.04 | 278.32 | 0.36 |
| 49 | MOL011604 | Miltirone II | 44.95 | 272.32 | 0.24 |
| 50 | MOL007155 | 3α-hydroxytanshinoneIIa | 44.93 | 310.37 | 0.44 |
| 51 | MOL007094 | Ginkgolide J | 44.84 | 424.44 | 0.74 |
| 52 | MOL007098 | Ginkgolide B | 44.38 | 424.44 | 0.73 |
| 53 | MOL007063 | 24-methylidenelophenol | 44.19 | 412.77 | 0.75 |
| 54 | MOL007122 | Physcion-8-O-beta-D-gentiobioside | 43.90 | 608.60 | 0.62 |
| 55 | MOL007059 | Poriferasterol | 43.83 | 412.77 | 0.76 |
| 56 | MOL007050 | Stigmasterol | 43.83 | 412.77 | 0.76 |
| 57 | MOL007058 | 24-ethylcholesta-5,22-dienol | 43.83 | 412.77 | 0.76 |
| 58 | MOL007049 | Fucosterol | 43.78 | 412.77 | 0.76 |
| 59 | MOL001495 | Dehydrotanshinone II A | 43.76 | 292.35 | 0.40 |
| 60 | MOL007108 | 6-Fluoroindole-7-Dehydrocholesterol | 43.73 | 402.70 | 0.72 |
| 61 | MOL000422 | Sclareol | 43.67 | 308.56 | 0.21 |
| 62 | MOL009620 | Bis[(2S)-2-ethylhexyl] benzene-1,2-dicarboxylate | 43.59 | 390.62 | 0.35 |
| 63 | MOL007105 | Salvianolic acid j | 43.38 | 538.49 | 0.72 |
| 64 | MOL000569 | Sitosterol alpha1 | 43.28 | 426.80 | 0.78 |
| 65 | MOL007150 | Przewaquinone E | 42.85 | 312.34 | 0.45 |
| 66 | MOL007149 | Tanshindiol B | 42.67 | 312.34 | 0.45 |
| 67 | MOL007068 | 4alpha,24-dimethylcholesta-7,24-dienol | 42.65 | 412.77 | 0.75 |
| 68 | MOL007051 | Obtusifoliol | 42.55 | 426.80 | 0.76 |
| 69 | MOL007036 | 24-methylenelanost-8-enol | 42.37 | 440.83 | 0.77 |
| 70 | MOL000006 | 4alpha-methyl-24-ethylcholesta-7,24-dienol | 42.30 | 426.80 | 0.78 |
| 71 | MOL007100 | 31-norlanosterol | 42.20 | 412.77 | 0.73 |
| 72 | MOL007085 | LAN | 42.12 | 426.80 | 0.75 |
| 73 | MOL009278 | (E,E)-1-ethyl octadeca-3,13-dienoate | 42.00 | 308.56 | 0.19 |
| 74 | MOL002883 | Mandenol | 42.00 | 308.56 | 0.19 |
| 75 | MOL002776 | Luteolin-4'-glucoside | 41.97 | 448.41 | 0.79 |
| 76 | MOL011588 | Kaempferol | 41.88 | 286.25 | 0.24 |
| 77 | MOL007151 | (6S,7R)-6,7-dihydroxy-1,6-dimethyl-8,9-dihydro-7H-naphtho[8,7-g]benzofuran-10,11-dione | 41.31 | 312.34 | 0.45 |
| 78 | MOL001558 | 2-isopropyl-8-methylphenanthrene-3,4-dione | 40.86 | 264.34 | 0.23 |
| 79 | MOL009644 | 7-O-Methylluteolin-6-C-beta-glucoside_qt | 40.77 | 318.30 | 0.30 |
| 80 | MOL001659 | Isogoycyrol | 40.36 | 366.39 | 0.83 |
| 81 | MOL007143 | Przewaquinone f | 40.31 | 312.34 | 0.46 |
| 82 | MOL007069 | Baicalin | 40.12 | 446.39 | 0.75 |
| 83 | MOL002651 | Cycloeucalenol | 39.73 | 426.80 | 0.79 |
| 84 | MOL009656 | Microstegiol | 39.61 | 298.46 | 0.28 |
| 85 | MOL007041 | α-amyrin | 39.51 | 426.80 | 0.76 |
| 86 | MOL001490 | Neocryptotanshinone ii | 39.46 | 270.35 | 0.23 |
| 87 | MOL007142 | Methyl (1R,4aS,7R,7aS)-4a,7-dihydroxy-7-methyl-1-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-1,5,6,7a-tetrahydrocyclopenta[d]pyran-4-carboxylate | 39.43 | 406.43 | 0.47 |
| 88 | MOL007048 | 4alpha,14alpha,24-trimethylcholesta-8,24-dienol | 38.91 | 426.80 | 0.76 |
| 89 | MOL007132 | Dan-shexinkum d | 38.88 | 336.41 | 0.55 |
| 90 | MOL000359 | Miltirone | 38.76 | 282.41 | 0.25 |
| 91 | MOL009615 | 1,2,5,6-tetrahydrotanshinone | 38.75 | 280.34 | 0.36 |
| 92 | MOL006824 | Cycloartenol | 38.69 | 426.80 | 0.78 |
| 93 | MOL009633 | Dihydrotanshinlactone | 38.68 | 266.31 | 0.32 |
| 94 | MOL001942 | 31-Norcyclolaudenol | 38.68 | 440.83 | 0.81 |
| 95 | MOL007124 | Lantadene A | 38.68 | 552.87 | 0.57 |
| 96 | MOL007107 | 31-norlanost-9(11)-enol | 38.35 | 414.79 | 0.72 |
| 97 | MOL007118 | Lophenol | 38.13 | 400.76 | 0.71 |
| 98 | MOL007079 | 24-methyl-31-norlanost-9(11)-enol | 38.00 | 428.82 | 0.75 |
| 99 | MOL007120 | CLR | 37.87 | 386.73 | 0.68 |
| 100 | MOL007082 | 4,24-methyllophenol | 37.83 | 414.79 | 0.75 |
| 101 | MOL007111 | Campesterol | 37.58 | 400.76 | 0.71 |
| 102 | MOL007115 | Campest-5-en-3beta-ol | 37.58 | 400.76 | 0.71 |
| 103 | MOL002222 | 24-methylenecycloartan-3beta,21-diol | 37.32 | 456.83 | 0.80 |
| 104 | MOL011594 | Genkwanin | 37.13 | 284.28 | 0.24 |
| 105 | MOL007121 | Przewalskin a | 37.11 | 398.49 | 0.65 |
| 106 | MOL007077 | 24-ethylcholest-22-enol | 37.09 | 414.79 | 0.75 |
| 107 | MOL009604 | Methylenetanshinquinone | 37.07 | 278.32 | 0.36 |
| 108 | MOL007081 | Poriferast-5-en-3beta-ol | 36.91 | 414.79 | 0.75 |
| 109 | MOL007070 | Beta-sitosterol | 36.91 | 414.79 | 0.75 |
| 110 | MOL007064 | Daucosterol_qt | 36.91 | 414.79 | 0.75 |
| 111 | MOL007071 | Sitosterol | 36.91 | 414.79 | 0.75 |
| 112 | MOL008400 | Syringetin | 36.82 | 346.31 | 0.37 |
| 113 | MOL010234 | Miltipolone | 36.56 | 300.43 | 0.37 |
| 114 | MOL002680 | Luteolin | 36.16 | 286.25 | 0.25 |
| 115 | MOL009660 | Sugiol | 36.11 | 300.48 | 0.28 |
| 116 | MOL011578 | C09092 | 36.07 | 286.50 | 0.25 |
| 117 | MOL009677 | Chryseriol | 35.85 | 300.28 | 0.27 |
| 118 | MOL009621 | Laricitrin | 35.38 | 332.28 | 0.34 |
| 119 | MOL009622 | 14b-pregnane | 34.78 | 288.57 | 0.34 |
| 120 | MOL000953 | 1-methyl-8,9-dihydro-7H-naphtho[5,6-g]benzofuran-6,10,11-trione | 34.72 | 280.29 | 0.37 |
| 121 | MOL009662 | NSC 122421 | 34.49 | 300.48 | 0.28 |
| 122 | MOL003044 | 4-methylenemiltirone | 34.35 | 266.36 | 0.23 |
| 123 | MOL007093 | Lanost-8-en-3beta-ol | 34.23 | 428.82 | 0.74 |
| 124 | MOL007088 | Lanost-8-enol | 34.23 | 428.82 | 0.74 |
| 125 | MOL009651 | 5,6-dihydroxy-7-isopropyl-1,1-dimethyl-2,3-dihydrophenanthren-4-one | 33.77 | 298.41 | 0.29 |
| 126 | MOL002881 | Salvilenone Ⅰ | 32.43 | 270.40 | 0.23 |
| 127 | MOL005043 | Ethyl oleate (NF) | 32.40 | 310.58 | 0.19 |
| 128 | MOL003578 | 3-beta-hydroxymethyllenetanshiquinone | 32.16 | 294.32 | 0.41 |
| 129 | MOL007140 | Delta-carotene | 31.80 | 536.96 | 0.55 |
| 130 | MOL011597 | Salviolone | 31.72 | 268.38 | 0.24 |
| 131 | MOL009665 | Diosmetin | 31.14 | 300.28 | 0.27 |
| 132 | MOL008173 | Salvilenone | 30.38 | 292.40 | 0.38 |
RALRG, Radix Salviae-Angelicae Sinensis Radix-Lycii Fructus-Rehmanniae Radix Praeparata-Ginkgo Folium; OB, oral bioavailability; MW, molecular weight; DL, drug-likeness.
Potential target genes and the PPI network map of RALRG
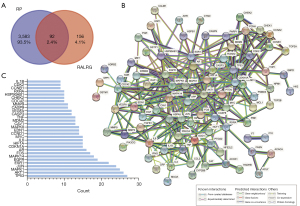
A total of 3,675 genes were obtained after the deduplication of RP-related genes downloaded from the GeneCards and OMIM databases. Among the 132 active ingredients, 248 target genes were identified. Using the Venn diagram, 92 intersection target genes were acquired (Figure 2A) and listed (Table S1). The PPI network was obtained using the STRING database, excluding disconnected nodes and with the highest score confidence (0.9000) (Figure 2B). The top 30 genes for the PPI network results were listed, including TP53, AKT1, MAPK1, JUN, ESR1, EGFR, MAPK14, FOS, AR, CDKN1A, HIF1A, and IL6, among others (Figure 2C).
GO and KEGG pathway enrichment analysis
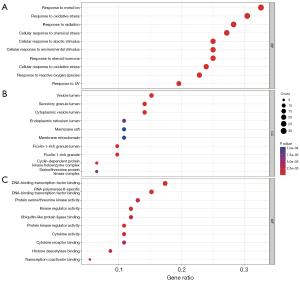
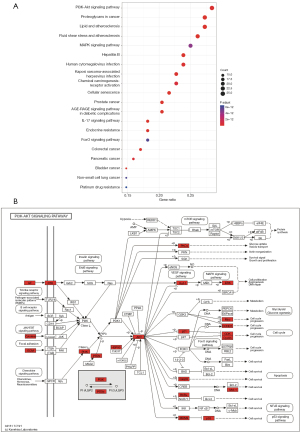
GO combines biological process (BP), cellular component (CC), and molecular function (MF), and the top 10 markedly enriched gene biological function catalogs were shown (Figure 3). BP enrichment showed that these target genes are mainly involved in oxidative stress, metal ion response, chemical stress, and so on (Figure 3A). CC enrichment showed that the target genes were primarily enriched in these subunits such as vesicle lumen, secretory granule lumen, cytoplasmic vesicle lumen, and ficolin-1-rich granule lumen (Figure 3B). MF enrichment showed that the target genes were mainly related to DNA-binding transcription factor binding, RNA polymerase II-specific DNA-binding transcription factor binding, kinase regulator activity, and protein kinase regulator activity (Figure 3C). These results suggested that the active ingredients in RALRG may treat RP through biological functions such as anti-oxidative stress. To further elucidate the mechanism of the active ingredients, a total of 162 signaling pathways (P adjust <0.05) were listed in the KEGG pathway enrichment analysis (Table S2). A bubble plot was created to show the top 20 pathways (Figure 4A). Many pathways were closely related to the occurrence and development of RP, such as the PI3K-AKT signaling pathway, cellular senescence, and MAPK signaling pathway. The essential PI3K-AKT signaling pathway is shown in Figure 4B.
Construction and analysis of the RALRG-RP-pathway potential pharmacology network
A visualized RALRG-RP-pathway potential pharmacology network (Figure 5) was constructed in Cytoscape 3.9.1 with 106 ingredients (excluding 26 ingredients without target genes) and 92 target genes, obtaining 195 nodes and 547 edges. In general, 1 gene was targeted by multiple active ingredients, and 1 ingredient targeted various genes. For example, Beta-sitosterol existed in Angelicae Sinensis Radix, Lycii Fructus, and Ginkgo Folium. Quercetin and luteolin existed in two herbs. A total of 23 pathways were mainly related to the RP mechanism (Table 2). The top 3 promising ingredients, including quercetin, luteolin, and kaempferol, were then selected by the MCC method, and their interaction with phenotype based on the experiment was concluded from the CTD (table available at https://cdn.amegroups.cn/static/public/atm-22-3557-1.xlsx). Quercetin, luteolin, and kaempferol were shown to protect astrocytes, microglial cells, RPE cells, and so on, via different BP.
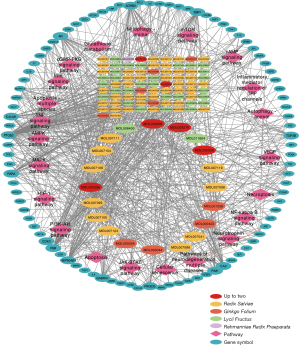
Table 2
| Description | P adjust | Count | Gene symbol |
|---|---|---|---|
| Cellular senescence | 6.22E−14 | 19 | CCNB1/CCND1/TGFB1/MYC/CCNA2/MAPK1/MDM2/CHEK2/CDK2/TP53/CDKN1A/IGFBP3/CALM1/PTEN/CHEK1/MAPK14/RAF1/IL6/AKT1 |
| PI3K-AKT signaling pathway | 3.51E−13 | 25 | MET/EGF/SPP1/CCND1/VEGFA/MYC/MAPK1/MDM2/EGFR/ERBB2/IL4/HSP90AB1/CDK2/TP53/CASP9/CDKN1A/PTEN/RXRA/RAF1/MCL1/IL6/KDR/PIK3CG/AKT1/PRKCA |
| MAPK signaling pathway | 4.48E−12 | 22 | HSPB1/MET/TNF/EGF/VEGFA/TGFB1/MYC/MAPK1/EGFR/ERBB2/MAPK8/TP53/IL1B/MAPK14/RAF1/CDC42/CASP3/KDR/FOS/AKT1/JUN/PRKCA |
| TNF signaling pathway | 7.30E−12 | 15 | EDN1/TNF/PTGS2/MAPK1/MAPK8/IL1B/CASP8/MAPK14/CCL2/IL6/CASP3/FOS/AKT1/JUN/MMP9 |
| VEGF signaling pathway | 1.99E−10 | 11 | HSPB1/PTGS2/VEGFA/MAPK1/CASP9/MAPK14/RAF1/CDC42/KDR/AKT1/PRKCA |
| HIF-1 signaling pathway | 7.66E−10 | 13 | EDN1/EGF/VEGFA/MAPK1/EGFR/ERBB2/CDKN1A/IL6/IFNG/HK2/HIF1A/AKT1/PRKCA |
| Apoptosis | 9.92E−10 | 14 | TNF/MAPK1/MAPK8/TP53/CASP9/CASP8/RAF1/CTSD/MCL1/CASP3/PARP1/FOS/AKT1/JUN |
| JAK-STAT signaling pathway | 5.31E−07 | 12 | EGF/CCND1/MYC/EGFR/IL4/CDKN1A/PIM1/RAF1/MCL1/IL6/IFNG/AKT1 |
| Pathways of neurodegeneration-multiple diseases | 7.41E−06 | 18 | PSMD3/TNF/PTGS2/MAPK1/SOD1/MAPK8/CASP9/CAT/IL1B/CALM1/CASP8/MAPK14/RAF1/IL6/CASP3/HSPA5/PRKCA/APP |
| Neurotrophin signaling pathway | 1.46E−05 | 9 | MAPK1/MAPK8/TP53/CALM1/MAPK14/RAF1/CDC42/AKT1/JUN |
| cAMP signaling pathway | 0.00031254 | 10 | EDN1/MAPK1/MAPK8/CALM1/ACOX1/RAF1/ADRB2/FOS/AKT1/JUN |
| Necroptosis | 0.00074059 | 8 | TNF/MAPK8/HSP90AB1/IL1B/CASP8/PARP1/IFNG/PYGM |
| Apoptosis—multiple species | 0.00087961 | 4 | MAPK8/CASP9/CASP8/CASP3 |
| Autophagy—animal | 0.00180027 | 7 | MAPK1/MAPK8/PTEN/RAF1/CTSD/HIF1A/AKT1 |
| Inflammatory mediator regulation of TRP channels | 0.00806022 | 5 | MAPK8/IL1B/CALM1/MAPK14/PRKCA |
| mTOR signaling pathway | 0.01259245 | 6 | TNF/MAPK1/PTEN/RAF1/AKT1/PRKCA |
| Mitophagy—animal | 0.01370082 | 4 | MAPK8/TP53/HIF1A/JUN |
| cGMP-PKG signaling pathway | 0.01651042 | 6 | MAPK1/CALM1/RAF1/ADRB2/PIK3CG/AKT1 |
| Wnt signaling pathway | 0.01651042 | 6 | CCND1/MYC/MAPK8/TP53/JUN/PRKCA |
| AMPK signaling pathway | 0.01683546 | 5 | CCND1/CCNA2/FASN/ACACA/AKT1 |
| Glutathione metabolism | 0.03973721 | 3 | GSTP1/GSTM1/G6PD |
| NF-kappa B signaling pathway | 0.04157345 | 4 | TNF/PTGS2/IL1B/PARP1 |
RP, retinitis pigmentosa.
Identification of key target genes and active ingredients
The PPI network was imported into Cytoscape for further analysis. The 10 highest-scoring genes were obtained using the MCC algorithm in the CytoHubba plugin, forming a sub-network (Figure 6A). A core sub-network consisting of 11 target genes was obtained using CytoNCA after filtering 3 times (Figure 6B-6D). Then, 9 target genes were acquired through the intersection of 2 core subnetworks (Figure 7A). The score and degree are shown in Figure 7B, and 9 targets were considered the hub genes. Therefore, the expression profiles of hub genes were presented by single-cell data (Figure 7C). JUN, which gained the highest score, was highly expressed in all cell types including bipolar cells, Müller cells, cone photoreceptor cells, rod photoreceptor cells, horizontal cells, and endothelial cells in the eye. Besides, the results of KEGG showed the PI3K-AKT signaling pathway was the most important, and AKT1 was shown to be expressed in different cell types in the retina. Therefore, transcription factor AP-1 (JUN) and RAC-alpha serine/threonine-protein kinase (AKT1) were selected as the most crucial protein for molecular docking. Next, molecular docking was performed to search and validate whether the top 3 promising ingredients and the targets, JUN and AKT1, have direct binding. Naturally, all three ligands could easily enter and bind to the active pocket of the selected proteins (Figure 8A-8F). The docking score is presented in Table 3. The interaction between three ingredients and the phenotype of cells based on the experiments were further concluded from the CTD (Figure 8G). In conclusion, these three ingredients may be potential complementary drugs and JUN and AKT1 may be the common targets for RP treatment.
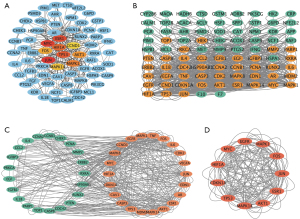
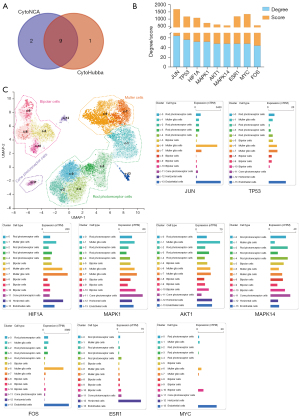
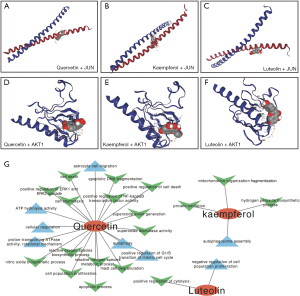
Table 3
| Target | Molecular name | Vina score | Cavity size | Center† | Size‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| X | Y | Z | X | Y | Z | |||||
| JUN | Luteolin | −6.1 | 354 | −25 | 17 | 19 | 21 | 21 | 21 | |
| Quercetin | −6.0 | 354 | −25 | 17 | 19 | 21 | 21 | 21 | ||
| Kaempferol | −5.8 | 110 | −15 | 18 | 28 | 21 | 21 | 21 | ||
| AKT1 | Luteolin | −6.1 | 110 | 17 | 5 | 14 | 21 | 21 | 21 | |
| Quercetin | −6.0 | 110 | 17 | 5 | 14 | 21 | 21 | 21 | ||
| Kaempferol | −5.9 | 102 | 16 | 16 | −2 | 21 | 21 | 21 | ||
†, docking pocket center coordinates; ‡, the size in the X, Y, and Z directions of the docking pocket. RP, retinitis pigmentosa.
Discussion
In this study, the RALRG-RP-pathway potential network revealed that RALRG had many ingredients acting on RP target genes, indicating that these herbs may be valuable for the therapy in RP and warrant further exploration. Radix Salviae is a traditional Chinese herb of which the dried roots are utilized for preventing neuronal apoptosis (7,22,23). Consistent with the enrichment results analysis, Behl concluded that its molecular mechanism involved anti-oxidation, inhibition of superoxide anion production in microglia, anti-inflammatory, and improvement of microcirculation (24). It also has protective effects on retinal tissue and vision by exhibiting severe Müller cell gliosis and enhancing dopamine D1 receptor function (25). The polysaccharide component of Lycii Fructus has been shown to have antioxidant, anti-inflammatory, anti-excitotoxic, and anti-apoptotic properties, making it a handy treatment option for the ocular environment (26). The results of several prospective randomized controlled clinical trials of Lycii Fructus in age-related macular degeneration (AMD) patients have shown that it could delay the progression of retinal degeneration and improve visual function (27,28). Montes et al. reported that Ginkgo biloba extract 761 had a protective effect on optic neurodegenerative diseases such as AMD, diabetic retinopathy (DR), retinal detachment (RD), glaucoma, and ischemic retinal diseases (29). Furthermore, diterpene ginkgolides meglumine injection, with extracts of Ginkgo biloba, was shown to inhibit apoptosis induced by optic nerve crush injury via modulating MAPK signaling pathways and inhibit p38, JNK, and ERK1/2 activation in retinal ganglion cells (30). In all, several mechanistic studies of these herbs can predict the efficacy of RALRG in treating RP, involving antioxidant, anti-inflammatory, anti-excitotoxic, and anti-apoptotic effects.
Although RP is a hereditary disease, non-genetic biological factors, including oxidative stress, also modulate or contribute to the disease progression (31). Excess reactive oxygen species (ROS) causes cellular oxidative stress damage, leading to cell dysfunction, necrosis, apoptosis, or autophagic cell death (32). The use of antioxidants such as curcumin (33), taurododecoxycholic acid (TUDCA) (34), n-acetylcysteine (35), lutein (36), and chlorogenic acid (37) can temporarily improve vision in PR patients. Besides, a recent study suggested that genetic mutations and overload of intracellular metal ions may cause deficiency or excess of metal ions, including Fe, Cu, and Zn, disrupting retinal metal homeostasis (38). The above results are consistent with our GO enrichment results, suggesting that RALRG can exert protective effects through multiple biological functions.
The PI3K/AKT signaling pathway is abnormally activated in many eye diseases, regulating the occurrence and development of eye diseases, consistent with the KEGG results in this study. PI3K/AKT signaling pathway is related to cell survival, metabolism, cell proliferation, angiogenesis, DNA repair, cell cycle progression, and glucose uptake vesicle transport (39). The ability of AKT to promote cell survival depends on its activation by PI3K. Once activated, AKT is involved in inactivating several pro-apoptotic proteins (39). Li et al. reported that human RPE cells could be protected from H2O2-induced oxidative stress by interfering with the upregulation of enzymes involved in the pro-survival PI3K/AKT pathway, suggesting that activating PI3K/AKT is a promising target in therapy (40).
Luteolin, quercetin, and kaempferol were identified as the most promising therapeutic ingredients for RP. Previous studies have shown that these three ingredients exert some protective effect on retinal structures and function by different mechanisms (41-44). Specifically, Liu et al. revealed that daily intraperitoneal injection of luteolin in rd10 mice at 100 mg/kg increased photoreceptor survival and improved retinal structure by inhibiting the JNK pathway to regulate retinal oxidative stress and inflammation (41). Ortega et al. revealed that quercetin treatment can stabilize the pathogenic rhodopsin mutant, inhibit ROS generation, attenuate microglial activation, and slow down photoreceptor degeneration (42). This indicates that quercetin is a promising complementary therapy. Studies have also shown that kaempferol could inhibit cell migration by targeting ERK1/2 signaling in human RPE cells and protect against oxidative stressed-human RPE cell damage through its antioxidant activity and antiapoptotic function (43,44). Our study found that kaempferol had good binding sites with both JUN and AKT1 protein, indicating that it may act on RP through the PI3K/AKT pathway to play a specific protective role. In conclusion, previous reports support that these three ingredients aforementioned may be promising potential drugs for RP treatment.
Key target genes may be promising targets for the comprehensive treatment of RP patients regardless of genetic mutations present in RP. Specifically, JUN is dispensable for developmental cell death and axogenesis in the retina (45) and involves in many pathways (Table 2). HIF-1A stabilization has been shown to reduce retinal degeneration by providing a temporal neuroprotective effect on photoreceptor cell survival, glial activation, and antioxidant response at the early stages of RP (46). C-fos was shown to be essential for light-induced apoptosis of photoreceptors and was notably continuously upregulated concomitant with apoptotic photoreceptor death RP (47). Consistent in our study, 9 key targets may play an important role in the mechanism of RALRG in treating RP.
However, we have only theoretically analyzed the mechanism of RALRG; experimental evidence is lacking. In future research, it is necessary to further verify and compare the effect of these ingredients in treating RP with the existing drugs.
Conclusions
The network pharmacological strategy integrates molecular docking to unravel the molecular mechanism of RALRG. Luteolin, quercetin, and kaempferol may be promising ingredients of RALRG for RP treatment. We believe that these findings may provide evidence for further drug discovery and development of RP.
Acknowledgments
We thank all the researchers for sharing data in public.
Funding: This study was funded by a grant from
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3557/rc
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3557/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3557/coif). All authors report the funding from National Natural Science Foundation of China (No. 81470625). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006;368:1795-809. [Crossref] [PubMed]
- Dias MF, Joo K, Kemp JA, et al. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog Retin Eye Res 2018;63:107-31. [Crossref] [PubMed]
- Verbakel SK, van Huet RAC, Boon CJF, et al. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res 2018;66:157-86. [Crossref] [PubMed]
- Russell S, Bennett J, Wellman JA, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 2017;390:849-60. [Crossref] [PubMed]
- Yamanaka S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell 2020;27:523-31. [Crossref] [PubMed]
- Xu J, Peng Q. Retinitis Pigmentosa Treatment with Western Medicine and Traditional Chinese Medicine Therapies. J Ophthalmol 2015;2015:421269. [Crossref] [PubMed]
- Ou C, Jiang P, Tian Y, et al. Fructus Lycii and Salvia miltiorrhiza Bunge extract alleviate retinitis pigmentosa through Nrf2/HO-1 signaling pathway. J Ethnopharmacol 2021;273:113993. [Crossref] [PubMed]
- Chudhary M, Zhang C, Song S, et al. Ginkgo biloba delays light-induced photoreceptor degeneration through antioxidant and antiapoptotic properties. Exp Ther Med 2021;21:576. [Crossref] [PubMed]
- Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:14. [Crossref] [PubMed]
- Hao da C. Xiao PG. Network pharmacology: a Rosetta Stone for traditional Chinese medicine. Drug Dev Res 2014;75:299-312. [Crossref] [PubMed]
- Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci 2019;20:4331. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 2021;49:D1388-95. [Crossref] [PubMed]
- Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. [Crossref] [PubMed]
- Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789-98. [Crossref] [PubMed]
- Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074-82. [Crossref] [PubMed]
- UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 2021;49:D480-9. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605-12. [Crossref] [PubMed]
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72. [Crossref] [PubMed]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235-42. [Crossref] [PubMed]
- Liu Y, Grimm M, Dai WT, et al. CB-Dock: a web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol Sin 2020;41:138-44. [Crossref] [PubMed]
- Lin TH, Hsieh CL. Pharmacological effects of Salvia miltiorrhiza (Danshen) on cerebral infarction. Chin Med 2010;5:22. [Crossref] [PubMed]
- Zhang Q, Xiao X, Zheng J, et al. Compound Danshen Dripping Pill Inhibits Retina Cell Apoptosis in Diabetic Rats. Front Physiol 2018;9:1501. [Crossref] [PubMed]
- Behl T, Kotwani A. Chinese herbal drugs for the treatment of diabetic retinopathy. J Pharm Pharmacol 2017;69:223-35. [Crossref] [PubMed]
- Chen YW, Huang YP, Wu PC, et al. The Functional Vision Protection Effect of Danshensu via Dopamine D1 Receptors: In Vivo Study. Nutrients 2021;13:978. [Crossref] [PubMed]
- Manthey AL, Chiu K, So KF. Effects of Lycium barbarum on the Visual System. Int Rev Neurobiol 2017;135:1-27. [Crossref] [PubMed]
- Bucheli P, Vidal K, Shen L, et al. Goji berry effects on macular characteristics and plasma antioxidant levels. Optom Vis Sci 2011;88:257-62. [Crossref] [PubMed]
- Li S, Liu N, Lin L, et al. Macular pigment and serum zeaxanthin levels with Goji berry supplement in early age-related macular degeneration. Int J Ophthalmol 2018;11:970-5. [Crossref] [PubMed]
- Montes P, Ruiz-Sanchez E, Rojas C, et al. Ginkgo biloba Extract 761: A Review of Basic Studies and Potential Clinical Use in Psychiatric Disorders. CNS Neurol Disord Drug Targets 2015;14:132-49. [Crossref] [PubMed]
- Fan XX, Cao ZY, Liu MX, et al. Diterpene Ginkgolides Meglumine Injection inhibits apoptosis induced by optic nerve crush injury via modulating MAPKs signaling pathways in retinal ganglion cells. J Ethnopharmacol 2021;279:114371. [Crossref] [PubMed]
- Murakami Y, Nakabeppu Y, Sonoda KH. Oxidative Stress and Microglial Response in Retinitis Pigmentosa. Int J Mol Sci 2020;21:7170. [Crossref] [PubMed]
- Gallenga CE, Lonardi M, Pacetti S, et al. Molecular Mechanisms Related to Oxidative Stress in Retinitis Pigmentosa. Antioxidants (Basel) 2021;10:848. [Crossref] [PubMed]
- Vasireddy V, Chavali VR, Joseph VT, et al. Rescue of photoreceptor degeneration by curcumin in transgenic rats with P23H rhodopsin mutation. PLoS One 2011;6:e21193. [Crossref] [PubMed]
- Fernández-Sánchez L, Lax P, Pinilla I, et al. Tauroursodeoxycholic acid prevents retinal degeneration in transgenic P23H rats. Invest Ophthalmol Vis Sci 2011;52:4998-5008. [Crossref] [PubMed]
- Lee SY, Usui S, Zafar AB, et al. N-Acetylcysteine promotes long-term survival of cones in a model of retinitis pigmentosa. J Cell Physiol 2011;226:1843-9. [Crossref] [PubMed]
- Berson EL, Rosner B, Sandberg MA, et al. Clinical trial of lutein in patients with retinitis pigmentosa receiving vitamin A. Arch Ophthalmol 2010;128:403-11. [Crossref] [PubMed]
- Shin JY, Yu HG. Chlorogenic acid supplementation improves multifocal electroretinography in patients with retinitis pigmentosa. J Korean Med Sci 2014;29:117-21. [Crossref] [PubMed]
- Liu S, Matsuo T, Miyaji M, et al. The Effect of Cyanine Dye NK-4 on Photoreceptor Degeneration in a Rat Model of Early-Stage Retinitis Pigmentosa. Pharmaceuticals (Basel) 2021;14:694. [Crossref] [PubMed]
- del Peso L, González-García M, Page C, et al. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 1997;278:687-9. [Crossref] [PubMed]
- Li Z, Dong X, Liu H, et al. Astaxanthin protects ARPE-19 cells from oxidative stress via upregulation of Nrf2-regulated phase II enzymes through activation of PI3K/Akt. Mol Vis 2013;19:1656-66.
- Liu XB, Liu F, Liang YY, et al. Luteolin delays photoreceptor degeneration in a mouse model of retinitis pigmentosa. Neural Regen Res 2021;16:2109-20. [Crossref] [PubMed]
- Ortega JT, Jastrzebska B. Neuroinflammation as a Therapeutic Target in Retinitis Pigmentosa and Quercetin as Its Potential Modulator. Pharmaceutics 2021;13:1935. [Crossref] [PubMed]
- Chien HW, Wang K, Chang YY, et al. Kaempferol suppresses cell migration through the activation of the ERK signaling pathways in ARPE-19 cells. Environ Toxicol 2019;34:312-8. [Crossref] [PubMed]
- Du W, An Y, He X, et al. Protection of Kaempferol on Oxidative Stress-Induced Retinal Pigment Epithelial Cell Damage. Oxid Med Cell Longev 2018;2018:1610751. [Crossref] [PubMed]
- Herzog KH, Chen SC, Morgan JI. c-jun Is dispensable for developmental cell death and axogenesis in the retina. J Neurosci 1999;19:4349-59. [Crossref] [PubMed]
- Olivares-González L, Martínez-Fernández de la Cámara C, Hervás D, et al. HIF-1α stabilization reduces retinal degeneration in a mouse model of retinitis pigmentosa. FASEB J 2018;32:2438-51. [Crossref] [PubMed]
- Hafezi F, Steinbach JP, Marti A, et al. The absence of c-fos prevents light-induced apoptotic cell death of photoreceptors in retinal degeneration in vivo. Nat Med 1997;3:346-9. [Crossref] [PubMed]


