Inertia based microfluidic capture and characterisation of circulating tumour cells for the diagnosis of lung cancer
Introduction
Identification of circulating tumour cells (CTC) in the blood of patients with cancer is now established in numerous cancers (1-3). The principals of cell capture is either using positive (e.g., cell size, antigen expression) (4-7) or negative (e.g., depletion of normal cells) selection (8) each having benefits and limitations. To date, the use of CTCs in routine clinical management has been limited to the current epithelial cell adhesion molecule (EpCAM)-based immunomagnetic approaches that only detect CTCs that express EpCAM, such as the CellSearch device (3). FDA approval for the CellSearch currently exists only for metastatic breast, prostate and colorectal cancer, thus excluding those CTCs with absent or low EpCAM expression as is the case with lung. Numerous reports have found low CTC recovery in non-epithelial and metastatic cancers such as melanoma, ovarian, pancreatic, and lung (9,10), underscoring the need for improved, unbiased CTC recovery technologies.
Once cells are captured, the next step is to characterise and correctly identify cancer cells, which has proved challenging in the context of generally low CTC loads (particularly in early cancer). There is also a lack of consensus of the diagnostic criteria used, and some of the criteria applied are not widely accepted e.g., E-cadherin or vimentin expression (11). Pathological criteria for diagnosis should be achieved amongst pathologists and professionals alike.
Recently the ClearBridge ClearCell FX systemTM (ClearBridge Biomedics, Singapore) was developed consisting of a spiral microfluidic device, separating and enriching CTCs from whole blood, based on cell size. Smaller haematological cells (red blood cells and leucocytes), 8–15 µm are affected by the Dean drag force and migrate to the outer wall, where larger CTCs (15–20 µm) are subjected to stronger inertial lift forces and focussed along the microchannel inner wall (12), resulting in a concentrated cell suspension.
This study evaluates the performance of the ClearBridge ClearCell FX systemTM to identify circulating lung cancer cells using conventional cytomorphological criteria assessed through haematoxylin and eosin (H&E) staining, and light microscopy, and define the test performance against an underlying diagnosis of cancer in patients with known or suspected lung cancer.
Methods
The study was carried out at the Royal Brompton & Harefield NHS Foundation Trust. The project was approved under the auspices of the National Institute of Health Biomedical Research Unit Advanced Disease Biobank (NRES 10/H0504/10).
A total of 52 participants were enrolled on the study, 36 of whom were patients with known or suspected lung cancer undergoing lung surgery, the remaining 16 consisted of healthy volunteers and non-cancer patients. All patients donated 7.5 mL of blood, one sample was excluded due to artefacts of blood processing, in total the final cohort consisted of 51 patients. The baseline characteristics of the patient are summarised in Table 1.
Full table
Peripheral blood was collected in 9 mL EDTA vacutainers or Streck tubes, either prior to surgery for surgical patients, and in clinics for all others, samples were processed within 24 hours for EDTA samples, and within 72 h for Streck tubes. The ClearBridge ClearCell FX systemTM (ClearBridge Biomedics) was used to capture and enrich CTCs according to the manufacturers protocol. Briefly, 7.5 mL of blood was mixed with 22.5 or 30 mL of red blood cell lysis buffer (G-Biosciences, St. Louis, MO, USA) at room temperature for 10 min. After incubation the samples were centrifuged at 500 g for 10 min followed by aspiration of supernatant, and finally addition of 4.3 mL of suspension reagent supplied by the manufacturer. The samples were then processed through the ClearCellTM system using a CTC ChipTM FR1 and microfluidics technology to separate tumour cells from white blood cells, to form an enriched suspension of CTCs. Processed samples were air dried on to glass slides and subjected to standard H&E staining, and then reported independently by two histopathologists. Results were categorised into “negative” or “positive” for cells suggestive of tumour, based on the presence of nucleated cells with high nuclear/cytoplasmic ratios that were larger than resting lymphocytes and/or cells having irregular nuclear outlines.
Inter-pathologist agreement was measured by Cohen’s kappa statistic and diagnostic utility was calculated as sensitivity, specificity, positive and negative predictive values and expressed with a 95% confidence interval. Statistical analyses were performed on Stata 13 (College Station, Texas, USA).
Results
From 30 July to 15 December 2015, blood specimens were processed from 51 participants with a mean age (SD) of 57 (15.04), 53% were male and primary lung cancer diagnosed in 24 (47%), secondary cancer diagnosed in 8 (16%) and no cancer (healthy control) in 19 (37%). Atypical cells suggestive of cancer were detected in an average of 49% of all cases, and 78.4% in all cancer cases. In early stage cancer this ranged from 50% and 66.7%, and late stage 0% and 100.0% for the first and second reporting team respectively. In both reporting groups, 9 (47.3%) of cases were reported as being suspicious of atypical cancer cells, despite being part of the benign cohort.
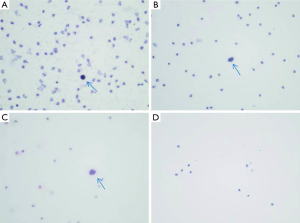
Based on the capture of CTCs and H&E staining (Figure 1), the diagnosis of cancer was reported for 28 (88%) and 22 (69%) of patients of the cancer group by each pathologist team respectively. The between pathologist agreement was 80% with a kappa (SE) of 0.61 (0.11; P<0.001) indicating substantial agreement according to Landis and Koch criteria (13).
Sensitivity, specificity, positive and negative predictive values were estimated for each pathologist team independently and were poor, as reported in Table 2.

Full table
Discussion
Various studies have shown the ability to isolate and capture CTCs in lung cancer patients, using a range of platforms, from antibody based (14,15) to filtration based techniques (4,5), with varying levels of performance and success.
The purpose of this study was to assess the ability of the ClearBridge ClearCell FX systemTM to isolate CTCs in the peripheral blood of surgical patients known or suspected of having lung cancer, using standard cytomorphologic criteria in a clinical setting.
Our investigation builds on the knowledge from previous studies, including studies by our group using a microfluidic biochip by ClearBridgeTM (4), and using another antibody independent filtration device, ScreenCellTM (5). Other similar studies for lung cancer include those using the device ISET, as well as the FDA approved antibody based platform, CellSearchTM (16). Although CellSearchTM results proved less efficient. Many groups took to comparing performances of differing devices, particularly in the context of antibody independent and antibody based, to enhance the argument of EpCAM loss and antibody bias, as is known to be the case in lung cancer (3,16,17).
We also report atypical cells with cytomorphological features suggestive of cancer identified within the ‘non cancer control’ group. Whilst these might represent true cancer cells in patients who may subsequently develop clinical features of cancer, they are more likely to be false positive results, with the cells representing other cell types, such as larger lymphoreticular cells. Further studies will be required using immunohistochemistry to help define this cell population more accurately and to refine the diagnosis of tumour cells when identified.
Currently focus has shifted to the development of circulating free nucleic acids (ctNA) monitoring and measuring techniques as an alternative to CTCs. ctNAs are more abundant in comparison, accounts for tumour heterogeneity unlike individual CTCs, and more importantly allows for real time mutation profiling and resistance development (18-20). It has been reported that several studies have shown correlations between mutations found in the tumour and blood in excess of 95% (21). Other studies evaluating their effectiveness have thus far reported positive results (20,22,23) suggesting a shift towards ctNAs as a more robust blood based biomarker, possibly in combination with CTCs.
The results of this study are not substantive to support clinical application of this microfluidic CTC capture platform, for CTC detection using standard H&E staining for clinical diagnosis in its current form. Sensitivity and specificity are generally quite poor at 59% and 41%, and 53% for both reporting teams, respectively, owing to either the analytic sensitivity of the method or the absence of CTCs in the samples, coupled with a poor recovery rate of approximately 40%. The results also indicate a level of subjectivity amongst the pathologists which is reflected in the inter observer agreement of 80.4%. Despite a significant kappa statistic of 0.61 (P<0.001) between the reporting groups, there is clearly some ambiguity and subjectivity in the diagnostic criteria, particularly in the context of standard H&E staining and, as discussed above, application of immunohistochemistry will need to developed in this low cellularity environment.
The implementation of a robust CTC isolation platform for CTC quantification in to the clinical setting could have potentially multiple benefits as has been demonstrated, ranging from prognosis and diagnostics, to treatment monitoring (2,24,25). However, at current the test performance of the ClearBridge ClearCell FX systemTM device improvements are still required before clinical application, with further work to be undertaken with cell characterisation and more specific staining techniques to identify cancer.
Conclusions
The findings of our study suggest that a spiral microfluidic antibody independent platform to capture CTCs using standard H&E staining and light microscopy assessment has the potentials as a diagnostic blood test for lung cancer, however implementation in a clinical setting requires further validation.
Acknowledgements
The project was funded in part by ClearBridge Biomedics (who did not have any role in the interpretation of results, drafting or approval of the submitted manuscript that was under the sole discretion of the principal investigator and research team) and supported by the National Institute of Health Research Respiratory Disease Biomedical Research Unit at the Royal Brompton and Harefield NHS Foundation Trust and Imperial College London.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The project was approved under the auspices of the National Institute of Health Biomedical Research Unit Advanced Disease Biobank (NRES 10/H0504/10), and written informed consent was obtained from all patients.
References
- Karhade M, Hall C, Mishra P, et al. Circulating tumor cells in non-metastatic triple-negative breast cancer. Breast Cancer Res Treat 2014;147:325-33. [Crossref] [PubMed]
- Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 2014;7:1-11. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Lim E, Tay A, Von Der Thusen J, et al. Clinical results of microfluidic antibody-independent peripheral blood circulating tumor cell capture for the diagnosis of lung cancer. J Thorac Cardiovasc Surg 2014;147:1936-8. [Crossref] [PubMed]
- Freidin MB, Tay A, Freydina DV, et al. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer 2014;85:182-5. [Crossref] [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [Crossref] [PubMed]
- Hofman VJ, Ilie MI, Bonnetaud C, et al. Cytopathologic detection of circulating tumor cells using the isolation by size of epithelial tumor cell method: promises and pitfalls. Am J Clin Pathol 2011;135:146-56. [Crossref] [PubMed]
- Freydina DV, Chudasama D, Freidin MB, et al. Diagnostic Utility of Unbiased Circulating Tumour Cell Capture through Negative Depletion of Peripheral Blood Cells. Oncology 2015;89:360-4. [Crossref] [PubMed]
- Kurihara T, Itoi T, Sofuni A, et al. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J Hepatobiliary Pancreat Surg 2008;15:189-95. [Crossref] [PubMed]
- Gupta V, Jafferji I, Garza M, et al. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012;6:24133. [Crossref] [PubMed]
- Hou JM, Krebs M, Ward T, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol 2011;178:989-96. [Crossref] [PubMed]
- Hou HW, Warkiani ME, Khoo BL, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 2013;3:1259. [Crossref] [PubMed]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159-74. [Crossref] [PubMed]
- Wang S, Thomas A, Lee E, et al. Highly efficient and selective isolation of rare tumor cells using a microfluidic chip with wavy-herringbone micro-patterned surfaces. Analyst 2016;141:2228-37. [Crossref] [PubMed]
- Satelli A, Brownlee Z, Mitra A, et al. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule- and cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin Chem 2015;61:259-66. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. [Crossref] [PubMed]
- Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Freidin MB, Freydina DV, Leung M, et al. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem 2015;61:1299-304. [Crossref] [PubMed]
- Pietrasz D, Pécuchet N, Fabre E, et al. What future for circulating tumor DNA? Current data and prospects in colorectal, non-small cell lung and pancreatic cancers. Bull Cancer 2016;103:55-65. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Carter L, Metcalf R, Blackhall FH, et al. Biology and clinical relevance of circulating tumour cells. J Thorac Dis 2012;4:453-5. [PubMed]
- Cho EH. Circulating tumor cells as emerging tumor biomarkers in lung cancer. J Thorac Dis 2012;4:444-5. [PubMed]


