Protection against lupus-like inflammatory disease is in the LAP of non-canonical autophagy
Introduction
Immunological diseases have been proposed to exist as a continuum, with innate immune-mediated autoinflammatory disease at one end of the spectrum and adaptive immune-mediated autoimmune disease at the other (1). Systemic lupus erythematosus (SLE) is often described as the quintessential autoimmune disease, with disease manifestations existing in multiple tissues throughout the body, particularly the skin, kidneys and brain. Phenotype variability among individual sufferers, as well as differences in disease severity also lead to specific diagnostic and therapeutic dilemmas. Symptoms associated with SLE include fever, chest pain, swollen lymph nodes, a red rash most commonly found on the face, as well as painful Raynaud’s phenomenon and swollen joints, with many patients also developing arthritis. This diversity in clinical features is due to a loss of control of self-tolerance of both the innate and adaptive immune systems (2). This array of clinical features is mirrored by the complexity of the many factors which may cause the disease, including genetic, environmental and hormonal. It is widely postulated that the root cause of SLE may involve an unknown environmental agent, triggering one’s existing genetic susceptibility and underlying immune system defects, with vitamin D deficiency as one such example (3). One certainty is that the adaptive immune response orchestrates the vast majority of SLE manifestations, by the presence of activated autoimmune T- and B- lymphocytes. However, the root cause of the disease remains elusive, and a diagnosis of SLE is typically confirmed once an individual is experiencing at least four of the eleven common signs of the disease (4,5). Of these eleven, a positive test for antinuclear antibody (ANA) is typically the most shared feature among patients.
Abnormalities in apoptosis, a form of programmed cell death in which damaged or ageing cells are intricately disposed of, are leading causes of SLE susceptibility. Many adverse stimuli can initiate a break in immune tolerance, by which healthy cells are destroyed and their nuclear contents, such as DNA and histones, become exposed and proceed to act as autoantigens. Moreover, certain abnormal immunological functions may (potentially) allow adaptive immune cells, such as T- and B- lymphocytes, to avoid cell death and remain in the circulation (6). Sensitised B lymphocytes produce antibodies against exposed autoantigens, such as double-stranded DNA (dsDNA), resulting in the formation of protein-antibody complexes, with the capacity to aggregate and damage blood vessels (7).
Dysfunctional autophagy in lupus
It is perhaps no surprise that certain shortcomings in our body’s capacity to clear these ‘dying’ cells are continually being proposed to be at the core of SLE pathogenesis (8). In a disease characterised by dysfunctional apoptosis (9) and immune activity against intracellular components (10-12), one process stands out as potentially providing a link between exposure of autoantigens and a break in immune tolerance. Autophagy is a term first coined by Christian de Duve, to describe an intracellular degradation system which seeks to maintain homeostasis by sequestering cytoplasmic constituents and delivering them to lysosomes (13). By achieving this, the autophagic process acts as a cell survival pathway to maintain healthy cells, by degrading damaged organelles and eliminating invading pathogens. Further roles of autophagy include cellular housekeeping, and operating as a component of cellular integrated stress responses. Therefore, the hypothesis that autophagy is dysfunctional or impaired in SLE seems very plausible, when autophagy’s role is to clear dying or dead cells and prevent potential intracellular autoantigens from being exposed to the immune system. And indeed, single-nucleotide polymorphisms (SNPs) have been associated with genetic susceptibility to SLE (14-16).
Autophagy was first identified in yeast, with homologues of autophagy-associated genes (ATGs) later being identified in humans (17). In terms of signalling regulation, the autophagy pathway is vast and complex, but mammalian target of rapamycin (mTOR) is often identified as the key player in balancing protein translation and autophagy. mTOR complex 1 (mTORC1) achieves this by operating as a negative regulator of the pathway. Further upstream, other key players in nutrient and energy detection, such as adenosine monophosphate activated kinase (AMPK), also operate as regulators of the pathway (18). When mTORC1 activity is inhibited, due to amino acid depletion for example, the autophagy pathway is activated. This process relies on a series of ubiquitin-like conjugation systems to maintain cellular homeostasis. The processing of targeted material for degradation relies on the formation of a double membrane vesicle, which emerges at a site known as the phagophore assembly site (PAS) in yeast. However, a similar site has yet to be identified in humans, with many investigators believing the site to be endoplasmic reticulum (ER) associated (19).
In humans, an initiation complex with UNC-like kinase 1 and 2 (Ulk1/2) at its core, is responsible for phagophore initiation, followed by a conjugation system stemming from Beclin-1, as the double membrane vesicle begins to elongate. As the phagophore transitions into an autophagosome, the membrane is enriched with microtubule-associated protein light chain 3 II (LC3-II), which is the central component of the next conjugation system. LC3-II is the lipidated form of LC3-I, with the conversion between these two forms often being questionably employed, via immunoblot, as a measure of complete autophagic flux (20). Finally, once sequestered components are ready to be delivered to the lysosome, fusion between the two membranes forms an autophagolysosome, with the final conjugation system utilising Atg12 and Atg5 to achieve this fusion (21) (Figure 1).
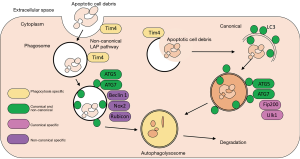
Canonical vs. non-canonical autophagy
Up until recently, there were considered to be three main forms of autophagy: micro, macro, and chaperone mediated. However, in 2011 a novel and alternate, or non-canonical, autophagy pathway was described (22,23). This distinct pathway and its variations, share many characteristics with the canonical pathway, but, its unique in that it operates independently of some of the complexes, which are integral to phagophore formation in normal autophagy. This alternative route is known as LC3-associated phagocytosis (LAP) and it is characterised by a reduced requirement for hierarchical influence of the Atg proteins, a double membrane that does not have to emerge from a single source, and the fact that Atg proteins can be recruited to a distinct membrane, other than the phagophore (24). Although some integral canonical components are not required for LAP, this phagocytic process has been shown to rely on certain components, which are conversely known to be dispensable for canonical autophagy (Figure 1). Two such examples include Rubicon and, to a lesser extent, an isoform of NADPH oxidase, NOX2. Recruitment of Rubicon to LAPosomes is integral to the incorporation of downstream proteins in the system, while it also stabilises a NADPH oxidase 2 (NOX2)-containing complex, thereby allowing reactive oxygen species (ROS) to be produced (25). Critically for LAP, phosphatidylinositol 3-phosphate (PtdIns3P) and ROS are both necessary if LC3 is to be conjugated to LAPosomes, as well as also being crucial for subsequent fusion with LAMP1(+) lysosomes (25). It is important to state that both canonical and non-canonical autophagy occur downstream of phagocytosis of cellular debris or dying cells; autophagy is responsible for degradation of this engulfed material. The principle difference between canonical and non-canonical autophagy is the fact that non-canonical autophagy acts independently of the RI1CC1 pre-initiation complex.
When one considers some of the immunological processes that are dependent on autophagy, such as antigen presentation (26), lymphocyte development (27) and regulation of inflammation (28), in the setting of both autoimmune and autoinflammatory disorders, it becomes clear how re-establishing functional autophagy could be beneficial, in terms of suppression, or even full correction of the disease phenotype.
In the 05 May 2016 issue of Nature, Martinez et al. published a letter describing the effects of defective non-canonical autophagy in vivo. In this article, and with regard to SLE specifically, Martinez et al. showed that adverse consequences may arise when non-canonical autophagy is not fully operational, and that disease phenotypes may ensue. Ultimately, both canonical and non-canonical autophagy strive for the same end result in much the same manner, but with intricate variations at key branches of the canonical pathway. Thus, it is likely that LAP has evolved to operate as a cellular safety net for whenever canonical autophagy is bypassed, or fails. This LAP-driven process drives cells towards the desired healthy phenotype that cellular homeostasis requires, and away from unnecessary, and often detrimental, apoptotic phenotypes.
LAP deficiency causes a lupus-like disease
The experimental animals used in Martinez et al. were conditionally ablated for key components of the LAP pathway, including components of canonical autophagy or T-cell immunoglobulin mucin protein 4 (TIM4), which is involved in macrophage engulfment of dying cells. This conditional ablation used lysozyme-M to target the knockdown to macrophages, monocytes, neutrophils and conventional dendritic cells. LAP deficiency created an autoinflammatory lupus-like syndrome in mice defective in the LAP specific proteins Becn1, Nox2 and Rubcn, as well as proteins that are common between canonical and non-canonical autophagy such as Atg7 and Atg5. However, proteins exclusive to the canonical autophagy pathway, Fip200 and Ulk1, did not produce the said phenotype; Tim4 ablation has been previously shown to induce a lupus-like phenotype in mice (Figure 1). The autoinflammatory lupus-like phenotype was induced by injection of irradiated thymocytes undergoing apoptosis, and was characterised by low body weight, the presence of anti-dsDNA antibodies and anti-nuclear antigens, in addition to increased levels of autoantibodies associated with autoimmune and autoinflammatory disorders. A key manifestation of SLE is kidney pathology and the mice with LAP deficiency presented with renal damage, as measured by increased serum creatine, blood urea nitrogen and proteinuria. LAP-deficient mice also had a strong interferon signature, a classical feature of lupus.
LAP deficiency causes dysfunctional degradation, not engulfment of dying cells
The engulfment or phagocytosis of cellular debris and dying cells occurs upstream of the autophagy pathways. Martinez and colleagues have previously published in vitro data (22) describing how the LAP pathway orchestrates the degradation of phagocytosed material. In their 2011 article, bone marrow derived macrophages (BMDMs) from GFP-LC3+ mice were used in a phagocytosis assay to study activation of the LAP pathway and how disruption of LAP induces a pro-inflammatory response from BMDMs. Not only did a deficient LAP pathway induce a pro-inflammatory cytokine secretion profile in BMDMs, but it also downregulated anti-inflammatory cytokine secretion, contributing to a loss of immune tolerance. In the present study, Martinez and colleagues continued this work by observing the ability of in vivo monocytes, macrophages, granulocytes and dendritic cells to not only engulf dying cells but also to degrade them and produce, in response, an appropriate cytokine secretion profile. In order to measure the extent of engulfment and degradation, apoptotic ultraviolet-irradiated thymocytes, labelled with a red fluorescent marker (PKH26), were injected into wild-type (WT) mice as well as Cre+Atg7f/f, Fip200f/f, Rubcnf/f and Tim4f/f mice. Engulfment was observed using immunofluorescence of labelled thymocytes within the phagocytes. Engulfment of dying cells was equivalent in all canonical and non-canonical autophagy-deficient mice and WT mice, except for the phagocytosis-deficient Cre+Tim4f/f mice, which were unable to engulf dying thymocytes. To measure the degree of degradation of engulfed cells, GFP-tagged LC3 was used as a marker of lysosomal fusion and cargo degradation. The Cre+Atg7f/f and Cre+RUBCNf/ mice were each unable to convert LC3 to LC3-II and, so, were unable to clear dying thymocytes, as a result of these mice being LAP deficient. WT mice and Cre+Fip200f/f mice successfully degraded the engulfed cargo, which is evidence in support of non-canonical, LAP-dependent autophagy being largely responsible for degradation of engulfed cells. When LAP-mediated autophagy is deficient, engulfed cells may remain in the phagocyte without being cleared, despite maintaining a functional canonical autophagy pathway.
In view of the fact that phagocytes which are deficient in the LAP pathway can successfully engulf dying cells but cannot degrade them because of their dysfunctional non-canonical autophagy mechanism, Martinez and colleagues investigated whether in vivo LAP-deficient macrophages were more pro-inflammatory than their LAP-sufficient counterparts. LAP-deficient macrophages produced pro-inflammatory IL-1β, IL-6 and CXCL10 after thymocyte engulfment, whereas, in contrast, LAP-sufficient macrophages produced anti-inflammatory IL-10. This was mirrored in serum levels of IL-1β, IL-6 and CCL4 observed in LAP-deficient animals but not in the LAP-sufficient animals, which produced increased serum IL-10 levels.
As LAP-deficient mice were shown to produce a lupus-like phenotype, with defective clearance of dying, engulfed thymocytes, Martinez and colleagues set out to investigate whether persistent administration of dying, engulfed thymocytes in LAP-deficient mice would aggravate this phenotype. And indeed it did, with the development of an SLE-like disease, characterised by increased ANAs, anti-dsDNA autoantibodies, glomerular deposits of IgG and C1q as well as increased alanine aminotransferase (ALT) levels. The increased serum pro-inflammatory cytokine profiles seen in LAP-deficient mice, administered with persistent dying, engulfed thymocytes, corroborated previous observations.
Conclusions
By studying non-canonical autophagy, Martinez and colleagues have uncovered a pivotal pathway, capable of not only general cellular housekeeping and homeostasis, but also protecting phagocytes from generating an autoinflammatory disease phenotype. The LAP pathway is the bedrock of both canonical and non-canonical autophagy and, without it, these processes are unable to achieve their ultimate purpose, namely the degradation of dying cells. LAP deficiency causes phagocytes to enter a severely pro-inflammatory phenotype, when engulfed cargo is resistant to disposal via autophagy. This is in stark contrast to the anti-inflammatory phenotypes of LAP-sufficient cells, which are capable of successful degradation of engulfed cargo. This innate immune dysfunction exacerbates systemic inflammation, by precipitating a vicious circle of apoptosis, failure to clear engulfed dying cells and development of further apoptosis. The resultant exposure of intracellular autoantigens, associated with lupus, then ensues and contributes to the autoinflammatory lupus-like disease that the authors describe.
Going forward, the link between the LAP pathway and IL-10 production is an intriguing line of enquiry (investigation), with some reports of dysregulated IL-10 in SLE (29). If the autophagy process is successful it then skews phagocytes towards an anti-inflammatory, IL-10 producing cell capable of protecting an individual from lupus, which open up avenues for therapeutic intervention. However, IL-10 levels are often normal or even elevated in lupus, perhaps being sourced from adaptive immune cells (30).
Autophagy is thought to degrade and regulate some essential intracellular pro-inflammatory protein complexes, the inflammasomes. These ancient innate immune cytokine-processing complexes are not often associated with the development of autoimmunity, but are rather characteristic of autoinflammatory conditions at the other end of the immunological disease spectrum, such as hereditary periodic fevers (31). However, cellular stress (32), changes in cell metabolism (33) and neutrophil extracellular traps (NETs) have all been described as priming NLRP3 inflammasome activity, and failure to clear engulfed cellular material certainly fits into this criterion (34,35).
The discovery of the function of the LAP pathway and the organism’s reliance on it to prevent the development of lupus is a major step forward in our understanding of the initiation of lupus, as well diseases from the autoinflammatory end of the immunological disease spectrum. Certain autoinflammatory diseases have arisen due to defects in various genes independent of autophagy, which may cause indirect malfunction of non-canonical autophagy, for example cystic fibrosis (CF) or hyperimmunoglobulinemia D with periodic fever syndrome (HIDS). CF is a monogenic disease in which a chloride ion channel is mutated and defective. Autophagy has long been described as being dysfunctional in CF, but as of yet it is not understood how the CF mutation causes dysfunctional autophagy and how this relates to disease phenotype. HIDS is characterised by altered cellular metabolism and activation of the NLRP3 inflammasome (36). However, it has been shown that autophagy is dysfunctional in phagocytes from these patients and this demonstration of the importance of the LAP pathway may explain the severe pro-inflammatory phenotype associated with HIDS (37). The LAP pathway is clearly crucial for maintaining cellular homeostasis, and may prove to be an underlying disease protagonist spanning right across the immunological disease continuum.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- McGonagle D, McDermott MF. A Proposed Classification of the Immunological Diseases. PLoS Med 2006;3:e297. [Crossref] [PubMed]
- Nagy G, Koncz A, Perl A. T- and B-cell abnormalities in systemic lupus erythematosus. Crit Rev Immunol 2005;25:123-40. [Crossref] [PubMed]
- Schneider L, Dos Santos AS, Santos M, et al. Vitamin D and systemic lupus erythematosus: state of the art. Clin Rheumatol 2014;33:1033-8. [PubMed]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1997;40:1725. [Crossref] [PubMed]
- Petri M, Orbai AM, Alarcón GS, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum 2012;64:2677-86. [Crossref] [PubMed]
- La Cava A. The busy life of regulatory T cells in systemic lupus erythematosus. Discov Med 2009;8:13-7. [PubMed]
- Kanta H, Mohan C. Three checkpoints in lupus development: central tolerance in adaptive immunity, peripheral amplification by innate immunity and end-organ inflammation. Genes Immun 2009;10:390-6. [Crossref] [PubMed]
- Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol 1994;152:3685-92. [PubMed]
- Lorenz HM, Grünke M, Hieronymus T, et al. In vitro apoptosis and expression of apoptosis-related molecules in lymphocytes from patients with systemic lupus erythematosus and other autoimmune diseases. Arthritis Rheum 1997;40:306-17. [Crossref] [PubMed]
- Wu H, Zhao M, Tan L, et al. The key culprit in the pathogenesis of systemic lupus erythematosus: Aberrant DNA methylation. Autoimmun Rev 2016;15:684-9. [Crossref] [PubMed]
- Lindau D, Mussard J, Rabsteyn A, et al. TLR9 independent interferon α production by neutrophils on NETosis in response to circulating chromatin, a key lupus autoantigen. Ann Rheum Dis 2014;73:2199-207. [Crossref] [PubMed]
- Lau CM, Broughton C, Tabor AS, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med 2005;202:1171-7. [Crossref] [PubMed]
- Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med 2003;9:65-76. [PubMed]
- International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN), Harley JB, Alarcón-Riquelme ME, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008;40:204-10. [Crossref] [PubMed]
- Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 2009;41:1228-33. [Crossref] [PubMed]
- Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009;41:1234-7. [Crossref] [PubMed]
- Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy 2008;4:740-3. [Crossref] [PubMed]
- Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132-41. [Crossref] [PubMed]
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010;12:831-5. [Crossref] [PubMed]
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1-222.
- Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298-302. [Crossref] [PubMed]
- Martinez J, Almendinger J, Oberst A, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 2011;108:17396-401. [Crossref] [PubMed]
- Florey O, Kim SE, Sandoval CP, et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 2011;13:1335-43. [Crossref] [PubMed]
- Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol 2011;13:7-12. [PubMed]
- Martinez J, Malireddi RK, Lu Q, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 2015;17:893-906. [Crossref] [PubMed]
- Münz C. Autophagy proteins in antigen processing for presentation on MHC molecules. Immunol Rev 2016;272:17-27. [Crossref] [PubMed]
- Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev 2010;236:95-109. [Crossref] [PubMed]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011;469:323-35. [Crossref] [PubMed]
- Llorente L, Richaud-Patin Y, Couderc J, et al. Dysregulation of interleukin-10 production in relatives of patients with systemic lupus erythematosus. Arthritis Rheum 1997;40:1429-35. [Crossref] [PubMed]
- Houssiau FA, Lefebvre C, Vanden Berghe M, et al. Serum interleukin 10 titers in systemic lupus erythematosus reflect disease activity. Lupus 1995;4:393-5. [Crossref] [PubMed]
- Peckham D, Scambler T, Savic S, et al. The burgeoning field of innate immune-mediated disease and autoinflammation. J Pathol 2017;241:123-139. [PubMed]
- Carta S, Penco F, Lavieri R, et al. Cell stress increases ATP release in NLRP3 inflammasome-mediated autoinflammatory diseases, resulting in cytokine imbalance. Proc Natl Acad Sci U S A 2015;112:2835-40. [Crossref] [PubMed]
- Lee GS, Subramanian N, Kim AI, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 2012;492:123-7. [Crossref] [PubMed]
- Kahlenberg JM, Carmona-Rivera C, Smith CK, et al. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol 2013;190:1217-26. [Crossref] [PubMed]
- Kahlenberg JM, Kaplan MJ. The inflammasome and lupus: another innate immune mechanism contributing to disease pathogenesis? Curr Opin Rheumatol 2014;26:475-81. [Crossref] [PubMed]
- Park YH, Wood G, Kastner DL, et al. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol 2016;17:914-21. [Crossref] [PubMed]
- Jurczyluk J, Munoz MA, Skinner OP, et al. Mevalonate kinase deficiency leads to decreased prenylation of Rab GTPases. Immunol Cell Biol 2016;94:994-9. [Crossref] [PubMed]


