Potential biomarkers for radiosensitivity in head and neck cancers
Introduction
Head and neck cancer is the sixth most common cancer in the world with about 600,000 new cases annually, with the 5-year overall survival rates between 50–60%. Head and neck cancers comprise tumors of the oral cavity, larynx, pharynx, salivary glands, and nasal passages, with squamous cell carcinoma (SCC) being the most common histological type. Currently, the known risk factors for head and neck squamous cell carcinoma (HNSCC) in developed countries include smoking and alcohol consumption (NCI Surveillance, Epidemiology, and End Results Program, http://seer.cancer.gov/statfacts/html/laryn.html). These represent about 1% of all new malignancies in the United States and the cumulative cost of care for patients with head and neck cancer is estimated at over 4 billion dollars annually by the year 2020 (NCI Cancer Prevalence and Cost of Care Projections, (https://costprojections.cancer.gov/graph.php). This is in part due to the side effects related to standard therapies, which consist of surgery, chemotherapy, and/or radiation. Effective strategies to alleviate unwanted toxicities to improve patient quality of life and reduce costs are elusive.
From a biological perspective, the human epidermal growth factor receptor (EGFR) was initially shown to be a key factor in the aggressiveness of head and neck cancers and their response to radiotherapy (1). This led to the landmark study combining the EGFR inhibitor cetuximab with radiation, which showed an overall survival benefit compared to radiation alone (2). Since then, no biologic therapy against new targets has shown benefit for patients with head and neck cancer. However, in recent years, it has become evident that the human papilloma virus (HPV) is not only an important risk factor, particularly amongst non-smokers; but is also associated with radiosensitivity in oropharyngeal cancer. Nonetheless, its clinical utility in other types of HNSCC is controversial (3,4).
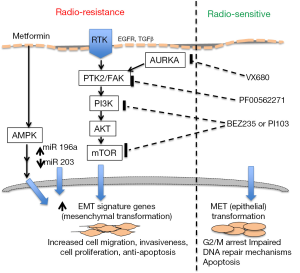
In this review, we discuss studies which report various biomarkers of radiation sensitivity in head and neck cancer and discuss the pathways which play important roles in the regulation of radiosensitivity. An overview of pathways is shown in Figure 1.
Biomarkers and radiosensitivity
Radiotherapy is one of the most important treatment modalities for various types of cancers including head and neck cancer. Disease-related mortality in HNSCC is due primarily to locoregional failure, thus understanding the mechanisms of radioresistance are imperative. Irradiation is known to affect various cellular processes to promote cellular damage and thus trigger tumor death. Some of those cellular mechanisms altered by irradiation include DNA repair, cell cycle regulation, and the reoxygenation of tumors. Technological advances have achieved improvements on targeting the irradiation precisely to the gross tumor and development of different fractionation modalities to optimize the biological effects of irradiation. Nonetheless, radiosensitivity still varies between tissues and tumor types.
Irradiation induces changes in gene expression in multiple cancer cell lines including HNSCC. In the genomic era, this type of comprehensive profiling has become more important in the pre-irradiation setting in order to identify molecular tumor signatures to predict radiosensitivity (Table 1). Identifying these biomarkers for various tumor types would permit a better stratification of patients based on their predicted response to irradiation. They would also increase the understanding of the intrinsic cellular mechanisms of radioresistance in cancer, thus promoting the development of adjuvant medications to personalize radiosensitization and improve patient outcomes.

Full table
To this end, the Torres Roca’s group created an algorithm to assign a radiosensitivity index (RSI) to tumors based on their genomic profile, in order to predict their response to irradiation (17-19). They identified a set of genes that were strongly associated to radiosensitivity and created RSI gene signature. Their findings in various types of cancers including rectal, esophageal, head and neck cancer, and breast cancer. Gene Ontology analysis showed that these genes were involved in DNA damage response, histone deacetylation, cell-cycle regulation, apoptosis, and proliferation; all of which play important roles in radiation response. The sensitivity and specificity of this molecular signature of radiosensitivity was high, and it did not correlate with response to other types of therapies.
Although the RSI has been validated in various cancer types, work from others suggests that each tumor behaves differently and thus genomic data cannot be extrapolated to all tumor types. In HNSCC, this aspect is supported by the previous findings of the positive-HPV status being associated to radiosensitivity only for oropharyngeal cancers compared to other HNSCC tumor sites. Given these limitations, the results from previous work in HNSCC (Table 1) are difficult to be interpreted particularly since there has been heterogeneity amongst the types and subtypes of the tumor tissues used, as well as the treatments received prior to analysis. Some studies have also suggested that a stem cell fraction within the tumors are the initiating pre-cancerous cells (20,21). These data highlight the importance of the context in which biomarkers are identified when designing signatures of radioresistance for HNSCC.
Recently, de Jong et al. [2015] reported a gene signature of radiosensitivity specific to laryngeal HNSCC (5). They used 32 HNSCC cell lines from primary laryngeal cancers with known radiosensitivity, which were not exposed to radiation or chemotherapy prior to collection. Their findings were then validated using patient tumor samples and genetically modified cell lines.
Statistically significant mRNA expression differences were found between cell line groups. They also observed an inverse correlation between microRNAs (miRs) relative to the cell lines’ radioresistance. miRs are short segments of non-coding single-stranded RNAs composed of 21-24 nucleotides that suppress gene expression by binding to complementary segments at the 3’-untranslated region of mRNAs. Since the discovery of miRs by Victor Ambros and Gary Ruvkun in 1993, there has been extensive work to characterize their specific targets and role in human disease (22,23). A role in cancer was first suggested by Calin et al. [2002] reporting a deletion of miR-15 and miR-16 in B-cell chronic lymphocytic leukemia (CLL) (24,25).
Twelve miRs were identified and linked to the regulation of the differentially identified mRNAs. These findings from their in vitro studies were then validated using samples from 34 pts with T2-3 tumors who were treated with radiotherapy alone with a curative intent (17 with local recurrence, 17 without local recurrence). The expression levels of the most significant miRs differentially expressed in the cell lines was tested in these patient tumor samples. Patients with local recurrence were found to have low miR-203 levels in their tumor samples. Although statistical significance was not achieved for other miRs, samples from patients with higher recurrence rates also showed lower expression of miR-452, miR-200b, and miR-141. The cellular functions associated with the differentially expressed mRNAs and miRs suggested epithelial-mesenchymal transition (EMT) mechanisms as predictive of radioresistance. However, allocation of the correct gene targets for every miR remains a challenge. Thus to validate that the differentially expressed mRNAs were involved in EMT, they used two sets of cell lines genetically modified to undergo EMT and their respective parental controls. The cells that had undergone EMT were more resistant to radiotherapy.
MicroRNAs and EMT
Multiple studies in cancer suggest that miRs play a role in cancer pathogenesis by their regulatory effects on EMT. EMT is a cellular developmental process associated with increased cellular mobility, migration, and invasion of cells. Thus EMT appears to be one important mechanism of tumor metastasis including in laryngeal cancer (26). Various pathways have been identified as potential targets to prevent EMT in cancer. Yang et al. [2016] recently proposed FAK/PI3K and AURKA as potential targetable pathways (27,28) (Figure 1).
In relation to the role of miRs in EMT, miR-200 family and miR-203 have been shown to be negative regulators of EMT (29). This appears to be mediated by a double negative feedback loop between Zeb1/2 and miR-200 family. Hypoxia and TGFβ upregulate the expression of Zeb1/2 which repress miR-200 gene expression thus initiating EMT. Conversely, increased levels of miR-200 inhibit Zeb1/2 and the cell status switches from mesenchymal to an epithelial state. The role of miR-203 in laryngeal cancer as suggested by de Jong’s findings are not surprising since this association with EMT has also been found in other cancers. Upregulation of Zeb1/2 and Snail1/2 suppress miR-203 in prostate and breast cancer cells, which then promotes EMT and tumor metastasis.
EMT and radiosensitivity
Recent work from Johansson et al. [2016] also supports a role of the EMT process in radioresistance (30). They found expression profiles of EMT signature genes, CDH1 (E-cadherin), CDH2 (N-cadherin), FOXC2, TWIST1, VIM, and FN1 in cell lines from 25 HNSCC primary tissues. Amongst the 25 cell lines, 4 showed a higher expression of the EMT signature genes. These four lines with EMT signature showed a mesenchymal morphology, higher migratory capacity, and radioresistance. It is also important to note that the 4 lines with highest EMT signature had a CD44 high/EGFR Low pattern previously associated with stemness in HNSCC and in mammary tumor cells.
The EMT signature was similarly observed in both the radioresistant cell lines and tumors from patients in the de Jong study, which includes a list of genes that also define cancer stem cells (31). Two cell lines that showed low expression of EMT genes were induced to undergo EMT via upregulation of the TGFβ pathway. Taken together, these data suggest that radioresistant cells exhibit EMT properties and biomarkers associated to EMT regulatory mechanisms.
Other pathways of radiosensitivity
One of the most common signaling pathways associated to radiosensitivity is the EGFR-PI3K/AKT pathway (32). In vitro exposure of prostate cancer cells to dual inhibitors of the PI3K/mTOR pathway triggered radiosensitization by shifting the cell cycle towards arrest in the G2 phase of the cell cycle. PI3K/mTor inhibition at G2 also caused a reduction in DNA double strand base repair and non-homologous end joining repair mechanisms, repressed colony formation, and induce apoptosis.
A recent relationship was reported between adenosine monophosphate-activated kinase (AMPK) and radioresistance in colon cancer samples (33). AMPK is a known regulator of cellular energy and reprogramming metabolism. Radioresistant tumors were found to have up-regulation of the AMPK protein and AMPK mRNA levels. On the contrary radiosensitive colon cancer cells showed down regulation of AMPK mRNA and protein levels. Further, activating the AMPK pathway in cancer cells with metformin promoted radioresistance in vitro, while inhibition of AMPK pathway by RNAi or chemical molecules re-sensitized radioresistant cancer cells. This study demonstrates the possibility of using AMPK pathway inhibitors as targeted therapies to enhance the radiosensitivity of tumors.
A proteomic and transcriptomic analysis of HPV-negative HNSCC was also recently performed that identified several proteins that were dysregulated in radioresistant cells including FGFR, ERK1, EGFR, and PTK2/FAK (34). In vitro inhibition of PTK2/FAK, but not FGFR, led to significant radiosensitization in several HNSCC cell lines. The mechanisms appeared to be potentiation of DNA damage by increased G2/M arrest of tumor cells. The PTK2/FAK protein expression was associated with its gene copy number, and correlated with outcomes on a cohort of HNSCC patients treated with radiation. A similar association was observed in the Head and Neck Cancer subgroup of the cancer genome atlas (TCGA). Thus, PTK2/FAK copy number could be a predictive genomic marker of radioresistance in HNSCC.
Summary
There is tremendous potential in this era of precision medicine to apply molecular signatures to predict the response of various tumors to radiotherapy. Many pathways are known to regulate radiation sensitivity, and novel biomarkers such as miRs are emerging to regulate such pathways. More evidence supports the EMT pathway in promoting radioresistance, and key players of EMT can be potentially targeted to enhance radiosensitivity of tumors including EGFR, TFGβ, mTOR, PKI3 and AMPK. These findings warrant further validation studies before implementing these signatures into the clinic.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Miyaguchi M, Olofsson JB, Hellquist HE. Expression of epidermal growth factor receptor in glottic carcinoma and its relation to recurrence after radiotherapy. Clin Otolaryngol Allied Sci 1991;16:466-9. [Crossref] [PubMed]
- Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med 2006;354:567-78. [Crossref] [PubMed]
- Morshed K. Association between human papillomavirus infection and laryngeal squamous cell carcinoma. J Med Virol 2010;82:1017-23. [Crossref] [PubMed]
- Young RJ, Urban D, Angel C, et al. Frequency and prognostic significance of p16(INK4A) protein overexpression and transcriptionally active human papillomavirus infection in laryngeal squamous cell carcinoma. Br J Cancer 2015;112:1098-104. [Crossref] [PubMed]
- de Jong MC, Jelle J, Grénman R, et al. Pretreatment microRNA expression impacting on epithelial-to-mesenchymal transition predicts intrinsic radiosensitivity in head and neck cancer cell lines and patients. Clin Cancer Res 2015;21:5630-8. [Crossref] [PubMed]
- Liu N, Boohaker RJ, Jiang C, et al. A radiosensitivity MiRNA signature validated by the TCGA database for head and neck squamous cell carcinomas. Oncotarget 2015;6:34649. [PubMed]
- Suh YE, Raulf N, Gäken J, et al. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. International journal of cancer. J Inter du Cancer 2015;137:1021. [Crossref]
- de Carvalho AC, Scapulatempo-Neto C, Maia DC, et al. Accuracy of microRNAs as markers for the detection of neck lymph node metastases in patients with head and neck squamous cell carcinoma. BMC Med 2015;13:108. [Crossref] [PubMed]
- Saad MA, Kuo SZ, Rahimy E, et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol Cancer 2015;14:181. [Crossref] [PubMed]
- Summerer I, Unger K, Braselmann H, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer 2015;113:76-82. [Crossref] [PubMed]
- Fukumoto I, Hanazawa T, Kinoshita T, et al. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer 2015;112:891-900. [Crossref] [PubMed]
- Darda L, Hakami F, Morgan R, et al. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PloS one 2015;10:e0122285. [Crossref] [PubMed]
- Wu Y, Yu J, Ma Y, et al. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol Lett 2016;12:871-8. [PubMed]
- Xu H, Yao Y, Meng F, et al. Predictive value of serum miR-10b, miR-29c, and miR-205 as promising biomarkers in esophageal squamous cell carcinoma screening. Medicine (Baltimore) 2015;94:e1558. [Crossref] [PubMed]
- Wong N, Khwaja SS, Baker CM, et al. Prognostic microRNA signatures derived from The Cancer Genome Atlas for head and neck squamous cell carcinomas. Cancer Med 2016;5:1619-28. [Crossref] [PubMed]
- Hilly O, Pillar N, Stern S, et al. Distinctive pattern of let-7 family microRNAs in aggressive carcinoma of the oral tongue in young patients. Oncol Lett 2016;12:1729-36. [PubMed]
- Torres-Roca JF, Eschrich S, Zhao H, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res 2005;65:7169-76. [Crossref] [PubMed]
- Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys 2009;75:489-96. [Crossref] [PubMed]
- Eschrich SA, Fulp WJ, Pawitan Y, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res 2012;18:5134-43. [Crossref] [PubMed]
- Shen LF, Zhou ML, Zhou SH, et al. Biomarkers of head and neck cancer stem cells and targeted therapeutic strategies. Int J Clin Exp Med 2016;9:614-25.
- Irani S. miRNAs Signature in Head and Neck Squamous Cell Carcinoma Metastasis: A Literature Review. J Dent (Shiraz) 2016;17:71-83. [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [Crossref] [PubMed]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993;75:855-62. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down- regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Yang L, Zhou Q, Chen X, et al. Activation of the FAK/PI3K pathway is crucial for AURKA-induced epithelial-mesenchymal transition in laryngeal cancer. Oncol Rep 2016;36:819-26. [PubMed]
- Wu J, Yang L, Shan Y, et al. AURKA promotes cell migration and invasion of head and neck squamous cell carcinoma through regulation of the AURKA/Akt/FAK signaling pathway. Oncol lett 2016;11:1889-94. [PubMed]
- Tang J, Li Y, Wang J, et al. Molecular mechanisms of microRNAs in regulating epithelial–mesenchymal transitions in human cancers. Cancer Lett 2016;371:301-13. [Crossref] [PubMed]
- Johansson AC, La Fleur L, Melissaridou S, et al. The relationship between EMT, CD44high /EGFRlow phenotype, and treatment response in head and neck cancer cell lines. J Oral Pathol Med 2016;45:640-6. [Crossref] [PubMed]
- Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 2005;115:1503-21. [Crossref] [PubMed]
- Chang L, Graham PH, Hao J, et al. PI3K/Akt/mTOR pathway inhibitors enhance radiosensitivity in radioresistant prostate cancer cells through inducing apoptosis, reducing autophagy, suppressing NHEJ and HR repair pathways. Cell Death Dis 2014;5:e1437. [Crossref] [PubMed]
- Jin H, Gao S, Guo H, et al. Re-sensitization of radiation resistant colorectal cancer cells to radiation through inhibition of AMPK pathway. Oncol lett 2016;11:3197-201. [PubMed]
- Skinner HD, Giri U, Yang L, et al. Proteomic Profiling Identifies PTK2/FAK as a Driver of Radioresistance in HPV-negative Head and Neck Cancer. Clin Cancer Res 2016;22:4643-50. [Crossref] [PubMed]


