Effective implementation of novel MET pharmacodynamic assays in translational studies
Introduction
MET oncogenic signaling (1,2) has been a promising drug target across many cancer types, albeit in a fraction of patients. So far, drugs that target MET tyrosine kinase (TK) have matched this promise only in preclinical models where MET is the driver gene. Among the first few candidate drugs that progressed to advanced stage testing, tivantinib and onartuzumab (Metmab), have failed to produce the desired results in phase III studies (3-5). This disappointment has led to questions regarding whether MET is a bona fide target for cancer therapy and also the value of rushing into untested patient selection strategies without fully establishing the linkage between the diagnostic test and the biology of MET dysregulation. Notwithstanding these setbacks, the pursuit of targeting MET is ongoing (6,7), as is evident from the >30 clinical trials registered at clinicaltrials.gov for testing new paradigms in targeting MET and from the clinical study of two new MET inhibitors this year. Success of these agents depends on improved patient selection strategies that we anticipate may overcome initial failures. Some recent developments are encouraging; most important among them is identification of exon 14 skipping mutations (MET∆14) in the TCGA data set, opening up an entirely new way and new patient population to identify and target MET dysregulation (8-10). Introduction of newer platforms for genomic interrogation such as circulating tumor DNA (ctDNA) in blood overcomes a major bottleneck (11,12) that is associated with the collection of fresh tumor biopsies. However, the approach of screening patients for MET∆14 and other genetic aberrations may be confounded by the biological diversity underlying these mutations. Therefore, establishing the relationship between MET mutations, as determined by ctDNA or other techniques, and dysregulation of the MET pathway in cancer tissue will be critical to ensuring the success of this new diagnostic strategy.
MET TK dysregulation happens in multiple ways, including abnormal ligand signaling, MET-activating mutations, and constitutive MET activation due to overexpression or gene amplification. Measurement of the phosphorylation state of key kinase domain tyrosines (broadly symbolized hereafter as pMET for phospho-MET) is the most reliable indicator of MET pathway activation, irrespective of its activation mechanism. Therefore, to demonstrate dysfunction of MET signaling or drug-induced suppression of MET signaling, pMET should be measured directly in tumor tissues. This has been achieved fairly consistently and successfully in preclinical models primarily using immunoblot techniques, and occasionally using ELISA or IHC assays. However, translation of matching assays in clinical settings and standardization of pre-analytic variables have been limited. In addition to difficulties in obtaining fresh tumor tissue to establish that MET diagnostics reflect the most recent status of cancer patients, two technical problems have hampered translation of pMET measurements in clinical studies: (I) sampling errors due to the limited stability of phospho-signals in biopsies; and (II) availability of specific antibodies and reagents that can be moved beyond the research setting into clinical laboratories. We recently described novel reagents and assays that the NCI has developed, standardized, and validated to facilitate the transition from the research lab to the clinic (13). In this review, we further elaborate on the underlying principles that guided development of our novel MET assays and the unique advantages they offer in providing tangible, quantifiable evidence of MET dysregulation and its blockade by MET inhibitors.
Ischemia time is the most critical pre-analytical variable for application of MET translational assays
For over a decade, measurement of phosphoproteins in vivo has been known to be influenced by delays in cryopreservation or fixation of tumor tissue (“time-to-stabilization”). This delay encompasses both warm ischemia (defined as the time between vessel ligation and tissue removal at body temperature) and cold ischemia (defined as the time between tissue removal and cryopreservation or fixation at room temperature). Despite recognition of the importance of this pre-analytical variable, standardization of this variable in clinical practice has lagged. For example, the most recent CAP-ASCO guideline (14) suggests that ischemia time should be minimized, not exceeding 60 minutes. However, multiple studies have proven that this timeline is inadequate if the goal is preservation of phospho-signals. Two recent proteomic studies employing MRM mass spectrometric assays for >20,000 serine (pS) and threonine (pT) phosphosites (representing 15,000 proteins) (15) and >200 tyrosine (pY) phosphosites revealed that >20% of pS or pT and >50% of pY signals can be altered or lost within 5 minutes of cold ischemia time (15,16). These studies could not identify MET peptides, likely because the enrichment process was insufficient to capture extremely low-abundance proteins. We studied the impact of both cold and warm ischemia time on SNU5 tumors and demonstrated that biopsies must be cryopreserved within three minutes of collection to preserve pMET signals (13). Even a modest delay of 15 minutes under warm/cold ischemia can result in >60% loss in pMET signal. Uncontrolled ischemia time will introduce measurement artefacts that may obscure results or simply lead to misinterpretation as drug-induced target suppression. Although a 3-minute collection time appears to be a daunting task, a coordinated effort between radiologists and laboratory personnel has enabled the successful collection of hundreds of biopsies within this time frame at NCI clinics. Details of biopsy collection and processing required for valid measurement of pMET and other PD biomarkers are described in SOP341205 at http://dctd.cancer.gov/ResearchResources/ResearchResources-biomarkers.htm. Fortunately, once cryopreserved, MET is fairly stable at assay operating temperatures in tissue lysates containing broad-spectrum phosphatase and protease inhibitors for the duration of analysis (13).
Tumor heterogeneity is another important pre-analytical variable that affects the consistency of MET measurements between biopsy cores. In experimental xenograft tumors, this variability dictates that drug-induced changes in pMET levels must exceed 45% in magnitude to be considered distinguishable from the variability due to normal biological heterogeneity and sampling error (13). There are many underlying reasons for this variability. Uneven sampling of tumor tissue may result in inconsistent tumor content, confounding the interpretation of assay results in pre- and post-drug treatment samples (17,18). When biopsies are converted to lysates this becomes an even greater challenge. We employed a total MET immunoassay that not only measures the full-length MET protein levels, but can also be used to normalize the measurement of MET phosphorylation. This allows for reporting of pMET signals as the ratio of pMET to total MET protein, potentially overcoming errors in tumor sampling (13).
MET-activating A-loop tyrosine-1235 is preferentially phosphorylated in some oncogenic forms
The biochemical evidence of MET activation and the mechanism of phosphorylation of tyrosines in the kinase domain have been extensively documented (2,19). In brief, ligand activation aggregates MET and disrupts the closed A-loop conformation, thus removing autoinhibition and allowing for kinase activation through trans-autophosphorylation of the A-loop tyrosine doublet, Y1234 and Y1235. Phosphorylation of both these tyrosines is essential for kinase activity of the wild type (WT) MET. The requirement for dual phosphorylation to maintain optimal kinase activity is similar to other kinases in this class and also represents an additional protection mechanism against deactivation by phosphatases. The sequence of phosphorylation of Y1234 vs. Y1235 during activation is not clearly established. It is proposed that Y1235 is more solvent-accessible and phosphorylated first (19). However, Y1235 phosphorylation is not enough to reverse the autoinhibition of WT MET, and Y1234 must also be phosphorylated for full kinase activity. In contrast to WT MET, many oncogenic mutations in the MET kinase domain (e.g., L1213V, M1268T, D1228H, D1228N, Y1235D, and M1250T) found in hereditary papillary renal carcinoma (HPRC) and sporadic renal tumors have been shown to make Y1234 phosphorylation dispensable in mutational studies (Figure 1A) (19,20). Oncogenic mutations in the MET gene can lower the threshold for activation by destabilizing the autoinhibited conformation when MET is only phosphorylated at Y1235 (Figure 1A). Despite these data establishing a critical role for Y1235, the measurement of pY1235-MET was not possible in the past as most antibodies recognize pY1234 or pY1235 with equal or unknown binding affinities (13). Therefore, for generation of specific monoclonal antibodies we prioritized Y1235 as the most critical phospho-tyrosine measurement in the MET A-loop. The goal was to develop an antibody that recognizes pY1235-MET independent of pY1234-MET. Development, specificity characterization, and application of a rabbit monoclonal antibody (clone 23111) specific to pY1235-MET—and not double-phosphorylated pY1234/35-MET—has been described recently (13). A pharmacodynamic assay employing this pY1235-MET-specific antibody has been developed and validated for clinical testing in human biopsies. This rabbit monoclonal antibody has also been converted to a recombinant antibody production system and is available for research use from the Developmental Studies Hybridoma Bank, University of Iowa (http://dshb.biology.uiowa.edu/ & https://antibodies.cancer.gov/apps/site/). The SOPs for assay methodologies of various MET assays are available from the NCI (http://dctd.cancer.gov/ResearchResources/ResearchResources-biomarkers.htm).
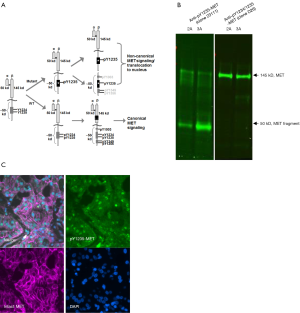
Accumulation of mono-phosphorylated Y1235-MET in hereditary papillary renal cancers
The first application of pY1235-MET antibody clone 23111 for human cancer tissues was performed on multiple tumor biopsies from an HPRC patient carrying germline trisomy of chromosome 7, with two copies of mutated MET (in this case, H1112R) and one copy of WT MET (20). We observed that pY1235-MET was mainly present as an approximately 50 kDa fragment (Figure 1B) by immunoblot and largely undetectable by sandwich assay using analyte capture at the N-terminal domain. Interestingly, the ~50 kDa fragment did not show reactivity with a dual pY1234/pY1235-MET antibody (clone D26), providing indirect evidence that this MET fragment lacks phosphorylation at Y1234 (however, the precise specificity of clone D26 has not been fully characterized). A quantitative immunofluorescence assay (qIFA) was developed for simultaneous detection of pY1235-MET and total MET in formalin-fixed paraffin-embedded (FFPE) tissues to determine localization of pY1235-MET or total MET signal in tissues and how it is modulated by MET inhibitors (21). The success of our qIFA was attributed mainly to greater specificity of our pY1235-MET antibody (clone 23111), as previously-existing antibodies crossreact not only with pY1234-MET, but also with other kinases such SRC and RON. Employing qIFA staining of formalin-fixed tumor tissues from an HPRC patient with the 23111 monoclonal antibody revealed that pY1235-MET staining was mostly localized to the nucleus (Figure 1C) (21).
Even though nuclear localization of a MET fragment has been described in multiple studies (22-24), our data (Figure 1C) for the first time demonstrates a uniquely phosphorylated form of MET in the cell nuclei of renal cancer patient tumor tissues. Our observations also confirmed earlier findings that a fraction of MET in sporadic renal tumors is primarily phosphorylated at Y1235 and not at Y1234 (based on non-reactivity with dual pY1234/pY1235-MET antibody). In light of these and prior findings, we speculate that the ~50 kDa fragment may be the gene product of the mutated MET gene copies, whereas the double-phosphorylated full-length MET detected is derived from the WT MET gene copy.
Our initial characterization provides sufficient evidence that our pY1235-MET antibody-based ELISA and qIFA assays can untangle the many diverse modes by which MET can be activated and processed. The specificity of clone 23111 was critical for measurement of phosphorylated MET in FFPE samples, where even minor crossreactivity renders most antibodies unreliable. Recent studies (25,26) highlight the importance of implementing a biomarker strategy that recognizes both canonical and non-canonical mechanisms of MET activation, recycling, and processing. Although most drugs act by suppressing the canonical pathway in MET-amplified cancer cell lines, non-canonical MET processing may be more prevalent in cancers with mutated MET, potentially predicting drug resistance and providing important information on targeting a subgroup of patients. Assaying for a truncated pY1235-MET could be an interesting ‘biomarker’ for HPRC, if it is proven that the mutant form mainly yields the uniquely phosphorylated ~50–60 kDa protein.
Nuclear localization may shed light on non-canonical functions of MET
Many receptor tyrosine kinases (RTKs) (e.g., EGFR, FGFR, VEGFR, ERRB2, and IGF1R) are known to translocate to the nucleus in response to ligand stimulation or oxidative stress to regulate transcription of genes (27,28), and similarly the role of MET in the nucleus is beginning to be elucidated.
Generally, nuclear transport occurs through a protease-dependent cleavage of the transmembrane domain of RTKs to release the cytoplasmic domain during endocytosis, which can then be translocated to other organelles including the nucleus. In this regard, a pH-dependent nuclear localization signal (NLS) has been identified in the juxtamembrane region (H1068–H1079) of MET encoded by exon 14, which translocates to the nucleus both in vitro and in vivo (29). MET fragment(s) in the nucleus are reportedly associated with increased expression of SOX and β-catenin (30) to promote gene transcription and the transition to a mesenchymal, drug-resistant phenotype. A second mechanism, similar to EGFR, suggests intact MET can be transported to the nucleus by integral trafficking from the endoplasmic reticulum (ER) to the nuclear envelop. It is believed that oxidative stress can induce this type of ER-to-nucleus trafficking mode. It has been shown recently that full-length MET can associate with PARP1 in the nucleus and phosphorylate PARP1 at Y907 in MDA-MB-231 cell lines. Phosphorylation of Y907-PARP1 modulated enzymatic activity to reduce sensitivity to a PARP inhibitor, which was overcome by a combination of PARP and MET inhibitors (25). This study provides compelling preclinical evidence of non-canonical MET signaling and how an understanding of such signaling may reveal new ways to suppress resistance and treat MET-driven cancers.
Identification of nuclear pY1235-MET in biopsies from an HPRC patient prompted us to look into other mutant forms of MET, especially in the H596 non-small cell lung cancer (NSCLC) cell line that harbors the MET exon 14 splice variant (9). Lacking the aforementioned juxtamembrane region, this mutant MET leads to decreased Cbl binding, prolonged protein stability, extended cell signaling upon HGF stimulation, and increased tumorigenicity (31). Our initial interest in the H596 cell line was for use in pharmacodynamic studies where MET is activated in vivo by paracrine signaling, and not by the MET amplification observed in many gastric cancers. We generated such a model through the use of human HGF knock-in (KI) mice (hemizygous hHGFki/− and homozygous hHGFki/ki; AVEO Pharmaceuticals and The Jackson Laboratory) bred (at Biological Testing Branch, NCI) to have homozygous SCID (Prkdcscid) background for xenograft studies. These KI mice produce significant stromal human HGF, detectable in blood, in place of murine HGF. H596 was the only cell line out of several tested in this mouse model to demonstrate detectable phosphorylation of MET in vivo, and the tumors grew relatively rapidly compared to H596 tumors in normal SCID mice (Figure 2A). When we applied our full length pY1235-MET and pY1356-MET assays to H596 xenografts developed in HGF KI mice, surprisingly we found less than 5% of MET was phosphorylated at Y1235. Our qIFA assay detected MET on the plasma membrane and pY1235-MET signals localized to the nucleus. Interestingly, the signal intensity of nuclear pY1235-MET increased in proportion to the HGF dose (normal SCID, hemizygous KI, homozygous KI; Figure 2B,C,D). Increased nuclear pY1235-MET also correlated with a significant increase in the mesenchymal phenotype (based on vimentin:E-cadherin ratios; Figure 2E). The HGF/MET pathway has previously been shown to be activated in small cell lung cancer (SCLC) in vivo, leading to increased expression of mesenchymal markers as well as tumorigenic and chemoresistant tumor phenotypes; specific inhibitors of MET reversed the resistant mesenchymal characteristics and enhanced chemosensitivity of SCLC tumors (32). Similarly, we observed increased expression of cancer stem cell factors such as Slug, Snail and SOX9 in E–M transitioning tumor tissues adjacent to HGF-containing stroma with parallel diminishment of E-cadherin expression and enhancement of vimentin expression (33), thus demonstrating EMT and changes in cancer stem cell biology induced by an HGF-secreting stroma and associated with enhanced pY1235-MET localization to the nucleus.
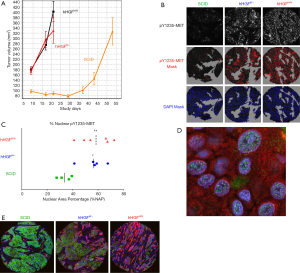
Phosphorylation at MET Y1234, Y1235, and Y1356 differs substantially in MET-amplified cell lines and tumor tissues
Several studies have suggested a relationship between phosphorylation of Y1234/1235-MET and A-loop activation, followed by phosphorylation of Y1349/1356-MET, which then regulates MET downstream signaling via multiple SH2 domain docking site proteins. However, the exact magnitude of phosphorylation at these tyrosine residues and their relation to the biological activity of MET is largely unknown. Most of our interpretation of the phosphorylation status of MET is derived from immunoblot data using antibodies that cannot distinguish between A-loop phosphorylated tyrosines or accurately reflect the relative quantitative changes in individual phosphorylated tyrosines. Thus, the relative proportion of total MET that is phosphorylated at Y1234 vs. Y1235 in cancer cells has not been studied. Absolute quantification requires MRM-MS-based quantitative assays in combination with stable isotope-labeled phosphopeptides as internal standards. We applied both MRM assays and ELISA, using our pY1235-MET specific antibody and recombinant MET as a calibrator, to determine pMET:MET ratios pre- and post-drug treatment. Our data, though limited by the number of cell lines, suggests interesting differences in the phosphorylation status of MET Y1234, Y1235, and Y1356. We observed that Y1235 is phosphorylated in significantly lower proportions as compared to Y1234. This was unexpected, as molecular-modeling-based predictions suggest Y1235 to be phosphorylated first, and most immunoblot data suggests equal phosphorylation of Y1234. We cannot rule out the possibility that our observations are influenced by the dephosphorylation rate of Y1234. In a gastric cancer model (SNU5), the pMET:MET ratio was 0.72 for pY1234/pY1235-MET, 0.21 for pY1235-MET alone, and 0.19 for pY1356-MET (Figure 3A,B). Our MRM-MS assays (34), developed to determine phosphorylation occupancy at Y1234 and Y1235 individually, revealed ratios similar to those measured by ELISA (Figure 3C). In multiple patient biopsies, where we could measure dual pY1234/35-MET by ELISA, the ratios of full-length pY1234/pY1235-MET:MET were 0.15, 0.28, and 0.38 in HPRC, ovarian, and esophageal cancers, respectively (35). These ratios were much lower than those observed in MET-amplified SNU5, MKN45, and GTL16 cell lines.
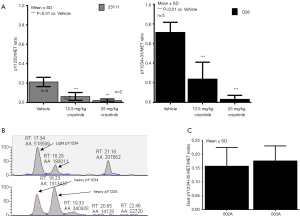
Interestingly, the degree of phosphorylation at the MET multiple-docking-site pY1356 is somewhat similar to pY1235. However, we could not confirm a direct relationship between these two sites, or whether the same MET molecule is activated at both sites (MRM assays for pY1356 were hampered by the solubility of tryptic peptides for calibration). Furthermore, despite similar phosphorylation of MET Y1235 and Y1356, suppression of Y1356 phosphorylation by small molecule MET inhibitors was significantly slower and lower in magnitude than that of Y1235 (13). These data for the first time highlight intricate differences in phosphorylation of different tyrosine residues on MET. Since it is the multiple-docking-site tyrosine that effects MET signaling, it is important to define the nuances of suppression of pY1356-MET in relation to the A-loop tyrosines and their molecular responses to drug exposure.
How does our ability to accurately measure pMET as a fraction of total MET help improve management of tumors driven by MET? An example is highlighted in a recent study describing a “rebound effect” during small molecule tyrosine kinase inhibitor (TKI) therapy (26). This study demonstrated that discontinuation of a drug leads to a small fraction of cells that remain alive and exhibit rebound activation of MET signaling. Importantly, this reactivation leads to a much “higher” level of phosphorylation than those seen in steady-state conditions. Furthermore, the “rebound” phosphorylation is prolonged in duration and accompanied by changes in MET processing and localization. Therefore, quantitative determinations of phosphorylation status pre- and post-treatment can provide important information on reactivation of MET that is linked to a resistance mechanism.
How to utilize pY1235-MET and pY1356-MET reagents/assays to support drug development
The pY1235-MET antibody can be used for diagnostic test
We have developed tumor sample collection and handling procedures that preserve the phospho-MET signal in biopsies. In addition, we have developed a specific antibody to pY1235-MET. Combined together, the pY1235-MET assays could not only be used in pharmacodynamic biomarker studies to demonstrate drug effects, but can also be used to identify patients with tumors that harbor constitutive pY1235-MET activation for treatment with MET inhibitors (a diagnostic test). Positive pY1235-MET staining on the membrane would confirm that MET is not only expressed but that signaling is “active”, indicating that both ligand-specific and MET inhibitors may be implemented in combination. On the other hand, nuclear staining would be informative for consequential MET processing and interaction with other pathways. This is a promising application that we have not yet validated in terms of demonstrating clinical utility with prospective tumor collection.
Link diagnostics to MET biology
The MET diagnostic biomarker landscape is changing drastically, with current efforts focused on genomic interrogation of MET to identify MET amplification and MET exon 14 alterations as potential predictive biomarkers. When combined with the ability to perform relatively non-invasive genomic tests in blood (ctDNA), this strategy supports effective screening of a large number of patients. Blood-based testing also enables a clinician to take a ‘snapshot’ of a patient’s current situation (cancer-associated gene mutations) before selecting a treatment regimen, capturing spontaneous mutations that may have occurred after a biopsy was taken, typically 2–3 months before treatment with a MET inhibitor begins. However, we believe that a blood-based diagnostic approach needs to integrate genetics with the cancer-specific biology of MET, at least in proof-of-concept studies. Otherwise we may be committing ourselves to the same unverified diagnostics that we know have not worked in the past. Despite more than two decades of anti-MET drug development, efforts to demonstrate suppression of MET phosphorylation and associated downstream signaling in clinical pre- and post-treatment biopsies are minimal. As such, there is a discrepancy between preclinical and clinical efficacy of MET inhibitors. It is critical to demonstrate direct evidence that not only is the diagnostic test identifying tumors harboring MET dysregulation, but also that drug-induced suppression is predictive of clinical responses. Since we are beginning a shift in diagnostic assessment of patients using new assays, it is critical to verify the linkage in early-stage clinical studies.
Use translational assays to validate hypotheses in proof-of-concept studies
Another unresolved issue is how best to combine MET inhibitors with other therapies that are predicted to activate MET signaling as a resistance mechanism. Multiple hypotheses have been proposed to combine MET-targeting therapy with cytotoxic agents, anti-angiogenesis inhibitors, agents targeting DNA repair, and immune therapies. We believe it is worthwhile to validate these hypotheses, which are largely based on in vitro data, by using MET translational assays to demonstrate that the mechanism of action is valid in vivo. We utilized our MET ELISA (13) and MET qIFA (21,33) assays to demonstrate MET activation as a possible resistance mechanism in response to VEGFR inhibition in a pre-clinical study. Our results confirmed the general hypothesis, but also highlighted important differences in responses depending on the epithelial or mesenchymal origin of the tumors. Thus, for future drug combination trials, there should be an emphasis on biomarker-driven proof-of-concept studies, clearly establishing the link between MET activation and a need to apply MET inhibitors in combination with other drugs. MET assays can also guide the exact time when MET inhibitors may be introduced in the combination study and the dosing schedules required to sustain MET suppression.
Identify biologically effective doses (BEDs) for highly selective MET inhibitors and avoid maximum tolerated dose (MTD)-based dosing
The phase II recommended dose has been traditionally selected on the basis of tolerability and usually is equivalent to the MTD defined by acceptable toxicity. However, many selective targeted agents may not reach MTD (36,37), or such doses may be significantly higher than those required for the optimal target suppression (36,38), also known as a BED. A recent review of cancer drug monotherapies found that >60% of approved drugs have doses lower than the MTD (36). For targeted therapies such as highly selective TKIs or monoclonal antibodies, BED provides a safe and effective alternative strategy. The biological effect could be determined by a pharmacodynamic biomarker assay in tumor tissues by establishing that marker suppression is predictive of tumor shrinkage. An example of this alternative approach is tepotinib (EMD1214063), which is a highly-selective small molecule MET inhibitor. Dose-limiting toxicities for tepotinib were not observed and a target engagement-based BED of 500 mg/day was recently selected for a phase II study (39). We applied pY1234/pY1235-MET assays as an “effect marker” to compare three MET inhibitors and showed that the dosage and schedules that achieve sustained suppression of pY1234/pY1235-MET >90% achieved optimal and equivalent tumor shrinkage irrespective of the type of MET inhibitor (40,41). Our results demonstrated that tepotinib achieved >90% suppression of pMET and >80% tumor regression in gastric tumor xenografts at a BID dose of 12.5 mg/kg (×5 days) (41). These data support the application of MET PD assays to guiding BED selection as an alternate strategy to MTD.
In conclusion, the newly developed MET assays provide robust tools for pharmacodynamic evaluation of MET inhibitors and could be utilized to improve our understanding of MET signaling in ways not previously feasible. The pY1235-MET-specific antibody could be applied to a new diagnostic test, as it is suitable for multiple testing platforms such as ELISA, immunoaffinity mass spectrometry, immunohistochemistry, and qIFA. However, the standards for demonstrating clinical utility of a diagnostic test are substantially different and require prospective clinical studies where the value of these reagents still needs to be proven.
Acknowledgements
We acknowledge Jennifer Weiner and Jeevan Prasad Govindharajulu for excellent technical work in the laboratory.
Funding: This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Disclaimer: The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89-103. [Crossref] [PubMed]
- Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov 2008;7:504-16. [Crossref] [PubMed]
- Rolfo C, Van Der Steen N, Pauwels P, et al. Onartuzumab in lung cancer: the fall of Icarus? Expert Rev Anticancer Ther 2015;15:487-9. [Crossref] [PubMed]
- Scagliotti G, von Pawel J, Novello S, et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol 2015;33:2667-74. [Crossref] [PubMed]
- Yoshioka H, Azuma K, Yamamoto N, et al. A randomized, double-blind, placebo-controlled, phase III trial of erlotinib with or without a c-Met inhibitor tivantinib (ARQ 197) in Asian patients with previously treated stage IIIB/IV nonsquamous nonsmall-cell lung cancer harboring wild-type epidermal growth factor receptor (ATTENTION study). Ann Oncol 2015;26:2066-72. [Crossref] [PubMed]
- Garber K. MET inhibitors start on road to recovery. Nat Rev Drug Discov 2014;13:563-5. [Crossref] [PubMed]
- Sheridan C. Genentech to salvage anti-MET antibody with subgroup analysis. Nat Biotechnol 2014;32:399-400. [Crossref] [PubMed]
- Awad MM. Impaired c-Met Receptor Degradation Mediated by MET Exon 14 Mutations in Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:879-81. [Crossref] [PubMed]
- Ma PC. MET receptor juxtamembrane exon 14 alternative spliced variant: novel cancer genomic predictive biomarker. Cancer Discov 2015;5:802-5. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Srivastava AK, Hollingshead MG, Weiner J, et al. Pharmacodynamic Response of the MET/HGF Receptor to Small-Molecule Tyrosine Kinase Inhibitors Examined with Validated, Fit-for-Clinic Immunoassays. Clin Cancer Res 2016;22:3683-94. [Crossref] [PubMed]
- Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997-4013. [Crossref] [PubMed]
- Mertins P, Yang F, Liu T, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics 2014;13:1690-704. [Crossref] [PubMed]
- Gajadhar AS, Johnson H, Slebos RJ, et al. Phosphotyrosine signaling analysis in human tumors is confounded by systemic ischemia-driven artifacts and intra-specimen heterogeneity. Cancer Res 2015;75:1495-503. [Crossref] [PubMed]
- Parchment RE, Doroshow JH. Theory and practice of clinical pharmacodynamics in oncology drug development. Semin Oncol 2016;43:427-35. [Crossref] [PubMed]
- Ferry-Galow KV, Makhlouf HR, Wilsker DF, et al. The root causes of pharmacodynamic assay failure. Semin Oncol 2016;43:484-91. [Crossref] [PubMed]
- Chiara F, Michieli P, Pugliese L, et al. Mutations in the met oncogene unveil a "dual switch" mechanism controlling tyrosine kinase activity. J Biol Chem 2003;278:29352-8. [Crossref] [PubMed]
- Pavlovich CP, Schmidt LS. Searching for the hereditary causes of renal-cell carcinoma. Nat Rev Cancer 2004;4:381-93. [Crossref] [PubMed]
- Navas T, Lawrence SL, Butcher D, et al. Quantitative immunofluorescence assessment of MET and epithelial to mesenchymal transition (EMT) biomarker modulation by antiangiogenic inhibitors in xenograft tumor tissues. Cancer Res 2014;74:abstr 1049.
- Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res 2006;66:7976-82. [Crossref] [PubMed]
- Gomes DA, Rodrigues MA, Leite MF, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem 2008;283:4344-51. [Crossref] [PubMed]
- Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis 2009;30:937-45. [Crossref] [PubMed]
- Du Y, Yamaguchi H, Wei Y, et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med 2016;22:194-201. [Crossref] [PubMed]
- Pupo E, Ducano N, Lupo B, et al. Rebound Effects Caused by Withdrawal of MET Kinase Inhibitor Are Quenched by a MET Therapeutic Antibody. Cancer Res 2016;76:5019-29. [Crossref] [PubMed]
- Chen MK, Hung MC. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J 2015;282:3693-721. [Crossref] [PubMed]
- Carpenter G, Liao HJ. Receptor tyrosine kinases in the nucleus. Cold Spring Harb Perspect Biol 2013;5:a008979. [Crossref] [PubMed]
- Chaudhary SC, Cho MG, Nguyen TT, et al. A putative pH-dependent nuclear localization signal in the juxtamembrane region of c-Met. Exp Mol Med 2014;46:e119. [Crossref] [PubMed]
- Xie Y, Lu W, Liu S, et al. Crosstalk between nuclear MET and SOX9/β-catenin correlates with castration-resistant prostate cancer. Mol Endocrinol 2014;28:1629-39. [Crossref] [PubMed]
- Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283-9. [Crossref] [PubMed]
- Cañadas I, Rojo F, Taus Á, et al. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clin Cancer Res 2014;20:938-50. [Crossref] [PubMed]
- Navas T, Pfister TD, Lawrence SM, et al. Impact of HGF knockin microenvironment on epithelial-mesenchymal transition and cancer stem cells in a non-small cell lung cancer xenograft model. Cancer Res 2015;75:abstr 5082.
- Meng Z, Srivastava AK, Zhou M, et al. Quantitation of Met tyrosine phosphorylation using MRM-MS. Methods Mol Biol 2013;1002:181-93. [Crossref] [PubMed]
- Srivastava AK, Govindharajulu JP, Park SR, et al. Pazopanib to suppress MET signaling in patients with refractory advanced solid tumors. J Clin Oncol 2016;34:abstr 2553.
- Sachs JR, Mayawala K, Gadamsetty S, et al. Optimal Dosing for Targeted Therapies in Oncology: Drug Development Cases Leading by Example. Clin Cancer Res 2016;22:1318-24. [Crossref] [PubMed]
- Postel-Vinay S, Arkenau HT, Olmos D, et al. Clinical benefit in Phase-I trials of novel molecularly targeted agents: does dose matter? Br J Cancer 2009;100:1373-8. [Crossref] [PubMed]
- Toska E, Baselga J. Pharmacology in the Era of Targeted Therapies: The Case of PI3K Inhibitors. Clin Cancer Res 2016;22:2099-101. [Crossref] [PubMed]
- Falchook GS, Kurzrock R, Amin HM, et al. Efficacy, safety, biomarkers, and phase II dose modeling in a phase I trial of the oral selective c-Met inhibitor tepotinib (MSC2156119J). J Clin Oncol 2015;33:abstr 2591.
- Srivastava AK, Hollingshead MG, Weiner J, et al. Application of MET pharmacodynamic assays to compare effectiveness of five MET inhibitors to engage target in tumor tissue. J Clin Oncol 2013;31:abstr 11103.
- Srivastava AK, Hollingshead M, Govindharajulu JP, et al. Met target inhibition-guided efficacy in preclinical models. Cancer Res 2014;74:abstr 3691.


