The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer?
Introduction
Lung cancer is the most common cause of cancer deaths (1-3). Non-small-cell lung cancer (NSCLC) represents of 85–90% of lung cancers (1-4). Epidermal growth factor receptor (EGFR) mutations occur in 10–30% of NSCLC (2,5-7). These mutations in the Caucasian population are present in about 10% (2). Identified for the mutation patients with advanced or metastatic NSCLC are candidates for personalized treatment. According to the international recommendations molecular testing should be carried out in patients with advanced NSCLC of a non-squamous subtype before first-line treatment starts (2). The researches indicated that those with tyrosine kinase inhibitors (TKIs)-sensitive EGFR mutations had longer progression-free survival (PFS) when receiving EGFR-targeted therapy (e.g., erlotinib) (8-11).
Hitherto, according to guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology published in 2013—“Pathologists should use formalin-fixed, paraffin embedded (FFPE) specimens or fresh, frozen, or alcohol-fixed specimens for polymerase chain reaction (PCR)—based EGFR mutation tests. Other tissue treatments should be avoided in specimens destined for EGFR testing” (3). But in the era of new technologies development it was easy to predict that this situation has to be changed. In the mid of 2016 the first liquid biopsy test was approved by the U.S. Food and Drug Administration (FDA) (12). In this paper the main meaning of novel technique of mutation detection in patients with NSCLC will be discussed briefly.
Liquid biopsy
Traditional biopsy is an invasive procedure to obtain a sample of the tumor tissue and is not always feasible. Liquid biopsy allows to identify patients whose tumors have specific mutations in the minimally invasive way and is less time-consuming. It is possible because tumor cells (and their DNA) are released into the circulation and circulating-free tumor DNA (cfDNA) could be isolated. Finally, the molecular test can be obtained. This is an alternative way to detect mutation and also an another way to gain a sample for testing in patients who previously could not be tested at all. Because of that it may play an important role in clinical decision making. It was previously reported that EGFR genetic testing was not being conducted in 19% of advanced NSCLC cases (13). The main reasons for not testing were: insufficient tissue, poor performance status (PS) and long turnaround time (13,14). Moreover, mutation test results were not available before treatment decision was taken in 23% of tested patients (13).
Because of the tumor heterogeneity a few samples should be tested to maximize genetic picture of the tumor. Nowadays, easily obtaining blood samples could serve also in this indication.
Finally, the easier way in which the liquid biopsy test is obtained also makes it useful in monitoring of disease progression. It is known that tumor cells evolve and often genetic changes are the reason for choosing new treatment. Thress et al. have demonstrated that analysis of cfDNA is able to detect mechanisms of resistance to EGFR-targeted therapies in NSCLC, e.g., EGFR T790M mutation (15). Since when osimertinib, an irreversible inhibitor of EGFR TKI-sensitizing and T790M resistance mutations is available, detection of this mutation is clinically meaningful (16).
The results of the study by Chabon et al. confirmed the utility of ctDNA-based resistance mechanism assessment, especially with detection of mutations present in multiple tumour deposits (17). Researchers employ CAPP-Seq (Cancer Personalized Profiling by deep sequencing) ctDNA analysis to study resistance mechanisms in NSCLC patients treated with the rociletinib (17). They observed a previously unrecognized high frequency of molecular heterogeneity (including a novel tertiary mutation in EGFR (L798I) and the emergence of activating KRAS mutations) in resistance mechanisms following treatment with EGFR TKIs (17). Interestingly, their findings suggest that pattern of resistance mechanisms to third-generation EGFR TKIs appear to be drug specific, and EGFR C797S mutations were noted in approximately 30% and 2% of patients treated with osimertinib and rociletinib, respectively (17). The usefulness of plasma ctDNA analyses were also shown in the study where T790M-positive patients treated with rociletinib upon progression were biopsied to explore rociletinib resistance (18). Moreover, Reckamp et al. have demonstrated that DNA derived from urine samples is feasible for mutation testing (19). The sensitivity of EGFR mutation detection in urine with tumor as a reference was 72% for T790M, 67% for exon 19 deletions, and 75% L858R mutations (for plasma samples: 93%, 87%, 100%, respectively) (19).
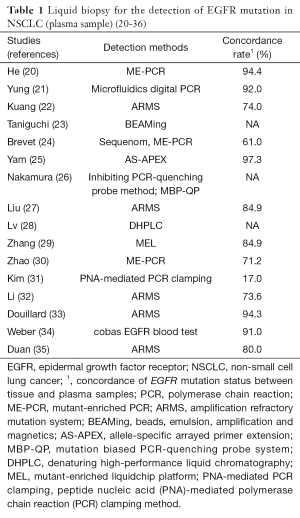
Hitherto, many techniques have been tested for the EGFR mutations detection using plasma samples. And several studies have demonstrated that mutations detected in plasma are highly concordant (usually 60–90%) with those detected in tumor tissue in NSCLC patients (Table 1) (20-36). For example, in the ASSESS study overall concordance of mutation status was 89% (14).
Full table
The concordance rate depends on method which was used (Table 1) (20-36). In general, digital genomic approaches (droplet digital PCR, BEAMing dPCR) are more sensitive than nondigital approaches (cobas and therascreen) (37). Moreover, in the AURA study, digital platforms appeared to detect a higher percentage of T790M mutations, compared with non-digital platforms (38). However, the cross-platform comparison showed that the cobas EGFR Mutation Test and BEAMing dPCR had highly concordant results, with high sensitivity (73–81%) for the detection of the T790M mutation and specificity ranging 58–67% (38).
While this method is an alternative way for mutation detection, we should remember that discordant genotypes between tumor biopsy and blood-based analyses were reported (39). And few things can influence on test results e.g., appropriate time of sample acquisition (cytotoxic agents may suppress the T790M ctDNA) (40).
The cobas EGFR Mutation Test
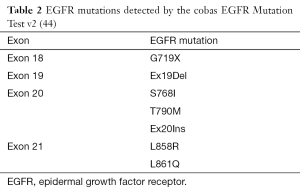
The cobas EGFR Mutation Test (v1) was approved on May 14, 2013 (41). This device is a real-time PCR test for the qualitative detection of exon 19 deletions and exon 21 (L858R) substitution mutations of the EGFR gene in DNA derived from FFPE human NSCLC tumor tissue (41). In 2015 the FDA approved cobas EGFR Mutation Test v2, adding e.g., T790M mutation to clinically important mutations, identified up to now by above-mentioned original test (42,43). Currently, this test detects EGFR mutations in NSCLC patients whose tumors have the exon 18 (G719X) substitutions, exon 19 deletions, exon 20 insertions and substitutions (T790M, S768I) and exon 21 substitutions (L858R, L861Q), but not any other EGFR mutations (Table 2) (44).
On June 1, 2016, FDA approved cobas EGFR Mutation Test v2 using plasma specimens as a companion diagnostic test for the detection of exon 19 deletions or exon 21 substitution mutations in the EGFR gene (12). This is the first liquid biopsy test approved for use by this agency. It allows for detection of mutations in cfDNA in less than 4 hours. If test is negative the routine testing using the FFPE tissue sample type is recommended (12).
The approval was based on a multicenter, open-label, randomised, Phase III ENSURE study, to evaluate the efficacy and safety of erlotinib versus gemcitabine plus cisplatin as first-line treatment for stage IIIB/IV NSCLC patients (8,12). Participants had tumour tissue specimens that tested positive for the EGFR exon 19 deletion or L858R mutations as determined by the cobas EGFR Mutation Test v1. Among patients screened and enrolled for this trial 86% and 98.6% had a plasma samples available for testing, respectively (12). Plasma was positive and negative for EGFR mutation in 76.7% and 98.2% of tissue-positive and tissue-negative cases, respectively (12).
Ongoing studies
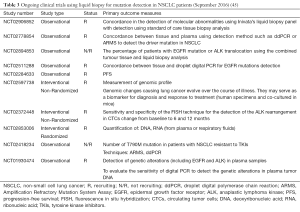
The usefulness of liquid biopsy is still widely tested in different settings (45). There are numerous studies that are in progress today and Table 3 shows a list of ongoing trials in the field of NSCLC (45). In the LIBIL study (NCT02511288), concordance (pourcentage) between tissue and droplet digital PCR for EGFR mutations detection and other molecular alterations routinely detected in NSCLC will be evaluated (45). Patients with stage IV EGFR-positive NSCLC may be enrolled into the NCT02284633 study. A biopsy and blood sample will be retrieved before treatment initiation. The patient will be monitored prospectively with blood samples every 3rd-6th week both during erlotinib treatment, subsequent lines of treatment and treatment intermissions. The blood samples will be analyzed for subtypes of EGFR M+ both sensitizing mutations and mutations known to drive resistance to erlotinib treatment. Patients will be followed until death or at least 24 months after inclusion. Any excess biological material will be stored for up to 15 years for future research purposes (Table 3) (45).
Full table
Interestingly, in the LEMA (Lung Cancer Early Molecular Assessment Trial) study, the potential usefulness of the early molecular profiling for all NSCLC patients, including stage I-III will be explored (45). This study is ongoing, but not recruiting participants (Last update: September 2016) (45).
Conclusions
Liquid biopsy is already a reality in clinical practice. A new era of molecular diagnostics is coming and plasma sample testing will also be used in other cancers and in other indications in the future. Researchers at The University of Texas MD Anderson Cancer Center have already published results using a PCR-based liquid-biopsy test called IdyllaTM BRAF Mutation Test which detects BRAF V600 mutations (46). They found that this test had 88–90% concordance with results from the standard tests (46). Also a pan-cancer diagnostic test being developed by Illumina is awaited (47). The development of cfDNA assays and their implementation into clinical practice is a valuable option in cases where tissue quantity is inadequate for mutation testing or in patients who refuse or are unable to undergo biopsy. But we have to be aware that the tissue biopsy and liquid biopsy do not compete. The liquid biopsy is another valuable option of mutation detection. The development of new molecular diagnostic tools allow more widely to use already approved targeted therapies and offer the right treatment to each patient in the best way possible.
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25:iii27-iii39. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasle MB, et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013;8:823-59. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Wu YL, Zhong WZ, Li LY, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: a meta-analysis based on updated individual patient data from the six medical centers in mainland China. J Thorac Oncol 2007;2:430-9. [Crossref] [PubMed]
- Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883-9. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Costa C, Molina-Vila M, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res 2014;20:2001-10. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- cobas EGFR Mutation Test v2. 2016. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm
- Spicer J, Tischer B, Peters M. EGFR mutation testing and oncologist treatment choice in advanced NSCLC: global trends and differences. Ann Oncol 2015;26:i57-i61. [Crossref]
- Reck M, Hagiwara K, Han B, et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: The ASSESS study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- FDA approves new pill to treat certain patients with non-small cell lung cancer. 2015. Available on: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm472525.htm
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature Communications 2016;7:11815. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- He C, Liu M, Zhou C, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 2009;125:2393-9. [Crossref] [PubMed]
- Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [Crossref] [PubMed]
- Kuang Y, Rogers BY, Yeap L, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotnib resistant on-small cell lung cancer. Clin Cancer Res 2009;15:2630-6. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino T, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 2011;73:96-102. [Crossref] [PubMed]
- Yam I, Lam DC, Chan K, et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol. 2012;7:1131-40. [Crossref] [PubMed]
- Nakamura T, Sueoka-Aragane N, Iwanaga K, et al. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J Thorac Oncol 2012;7:1369-81. [Crossref] [PubMed]
- Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. [Crossref] [PubMed]
- Lv C, Ma Y, Feng Q, et al. A pilot study: sequential gemcitabine/cisplatin and icotinib as induction therapy for stage IIB to IIIA non-small-cell lung adenocarcinoma. World J Surg Oncol 2013;11:96. [Crossref] [PubMed]
- Zhang H, Liu D, Li S, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn 2013;15:819-26. [Crossref] [PubMed]
- Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 2013;85:119-25. [Crossref] [PubMed]
- Kim HR, Lee SY, Hyun DS., et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 2013;32:50. [Crossref] [PubMed]
- Li X, Ren R, Ren S, et al. Peripheral blood for epidermal growth factor receptor mutation detection in non-small cell lung cancer patients. Transl Oncol 2014;7:341-8. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated Caucasian of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. [Crossref] [PubMed]
- Duan H, Lu J, Lu T, et al. Comparison of EGFR mutation status between plasma and tumor tissue in non-small cell lung cancer using the Scorpion ARMS method and the possible prognostic significance of plasma EGFR mutation status. Int J Clin Exp Pathol 2015;8:13136-45. [PubMed]
- Jiang T, Shengxiang R, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer 2015;90:128-34. [Crossref] [PubMed]
- Buder A, Tomuta C, Filipits M. The potential of liquid biopsies. Curr Opin Oncol 2016;28:130-4. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to suport the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Uchida J, Imamura F, Kukita Y, et al. Dynamics of circulating tumor DNA represented by the activating and resistant mutations in epidermal growth factor receptor tyrosine kinase inhibitor treatment. Cancer Sci 2016;107:353-8. [Crossref] [PubMed]
- cobas® EGFR Mutation Test–P120019. Available online: http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/DeviceApprovalsandClearances/Recently-ApprovedDevices/ucm352932.htm
- cobas® EGFR Mutation Test v2 - P150047. Available on: http://www.fda.gov/medicaldevices/productsandmedicalprocedures/deviceapprovalsandclearances/recently-approveddevices/ucm519922.htm
- Kwapisz D. ‘Signalling pathways in cancer’ — a report from the European Society for Medical Oncology symposium. Oncol Clin Pract 2016;12:63-6.
- cobas® EGFR Mutation Test v2. PMA P120019/S007: FDA Summary of Safety and Effectiveness Data. Available on: www.fda.gov
- Clinical Trials. Available on: https://clinicaltrials.go.v
- Janku F, Huang HJ, Claes B, et al. BRAF Mutation Testing in Cell-Free DNA from the Plasma of Patients with Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther 2016;15:1397-404. [Crossref] [PubMed]
- Illumina Forms New Company to Enable Early Cancer Detection via Blood-Based Screening. 2016. Available on: http://www.illumina.com/company/news-center/press-releases/press-release-details.html?newsid=2127903