Airway microbiome research: a modern perspective on surveillance cultures?
Ventilator-associated pneumonia (VAP)
The incidence of VAP is estimated to be around 10% in a high risk population of critically ill patients who are mechanically ventilated for more than 48 h (1). It is unclear if VAP is responsible for an increase in mortality (2), but it is strongly associated with an increase in ICU-length of stay, morbidity and healthcare costs (1,3). The optimal strategy to decrease the impact of VAP is three-folded: (I) prevent when possible; (II) early and targeted treatment when pneumonia develops; and (III) withholding antibiotics from those patients that develop symptoms but are not infected.
Over the last decade, major improvements have been made in the prevention of VAP, with great cost-effectiveness (4). Preventive measures have been targeted against the two pathophysiological principles of VAP development; micro-aspiration of bacteria that accumulate above the endotracheal tube (oral hygiene, semi-recumbent position, cuff pressure and suctioning of above the cuff) and the introduction of pathogens from the environment (hand hygiene, bacterial filters and minimal manipulation of the tubing). These pathophysiological principles have been relatively unchallenged over the last decades but fail to explain why patients who develop VAP and ventilator-associated trachea-bronchitis (VAT) carry the same pathogens (1). This suggests that there are either undiscovered pathogen factors that influence pathogenicity or that there are host or environmental factors that influence the development of VAP.
In this review, we will discuss recent advances in molecular biology, which allow for the culture-independent evaluation of the bacterial flora in the human body. Microbiome research in intubated and mechanically ventilated patients is in its infancy but has already shed light on several factors in the pathophysiology of VAP, which will be described in this review. A deepened understanding of the pathophysiology of VAP allows for better prevention and treatment. Furthermore, culture-independent techniques might allow for more targeted treatment of the causative pathogen in the near future.
Microbiome research
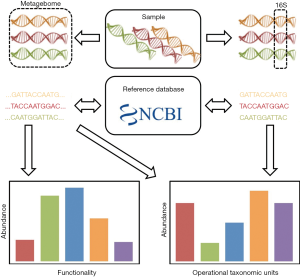
Microbiome research has been made possible through the development of high-throughput sequencing. Different types of analysis can be performed, depending on the deepness of nucleic acid sequencing. In general, there are two types of sequencing procedures (5). Metagenomics relies on the sequencing of all nucleic acids sequences obtained in a sample. This technique allows for full genome sequence, for instance of new species in a sample. In contrast, analysis of bacterial microbiota can also be performed based on certain conserved regions in the genome, which allows for the generation of a phylogenetic tree, which described the proportion of each bacterial species or phyla in a sample (Figure 1).
Phylogenetic sequences
Microbiota analysis is based on sequencing of specific regions of the DNA genes encoding the 16S ribosomal RNA (rRNA) subunit (6). This is made possible by the presence of regions that are conserved throughout the bacterial kingdom within these genes, which allows PCR amplification using universal primers. Eight hypervariable regions with sequences specific for each bacterial phyla and species separate these conserved regions. After next generation sequencing of the PCR amplified hypervariable regions, the sequence alignment on pre-identified sequence libraries permits the determination of bacterial composition in samples.
Strengths and limitations
A great advantage of sequencing techniques over conventional microbiological culture is its capability to identify hard-to-grow or even uncultivable bacteria. The 16S rRNA sequencing is also superior over conventional microbiology in terms of the possibility for sample storage as the DNA is stable when frozen, while culture dependent techniques rely on living bacteria and therefore fresh material. Several limitations should be understood in order to correctly interpret microbiota data. First, interpretation of 16S rRNA sequencing data relies on the number of hypervariable regions amplified and sequenced. Indeed, the precision of bacterial composition of a sample will vary depending on this number. In addition, the number of sequences obtained from a sample will determine the deepness of microbiota analysis. On the one hand, the greater hypervariable regions are sequenced and the greater number of sequences is obtained for a sample, the greater details are provided in term of bacterial species composition. On the other hand, the full length of 16S rRNA genes does not allow its complete sequencing for microbiota study in routine. Also, in order to reduce cost, many samples may be sequenced in the same run, reducing the number of sequenced obtained for each sample. Thus, a compromise has to be done between number of hypervariable regions, number of samples sequenced in the procedure, and financial and technical possibilities allowed for the study. Most recent studies also select one or two hypervariable regions and use the variation in those to generate phylogenetic trees. Based on these 16S rRNA sequences, the separation of bacterial species are poorly effective within some phyla, such as the Streptococcus (7).
Second, in contrast to conventional microbiology, microbiota analysis provides only proportional abundance of bacterial phyla and species and identifies both dead and live bacteria. Due to these characteristics, phylogenetic sequences do not allow for a quantitative measurement of the number of copies of specific bacterial species. This has to be integrated for data interpretation when repeated samples in a short period of time are performed (8,9).
Specificities for the lung
The lungs have a complex architectural structure from the trachea to the alveoli. Therefore, microbiota may not be homogenous throughout the lungs and this make interpretation of data somewhat difficult to compare. Various methods are available to obtain samples of the bacterial population of the airways. In ICU patients, the two most available technics are tracheal aspirations and bronchoalveolar lavages. However, the sampling methods by themselves may induce a contamination from proximal parts such as the oropharyngeal or oronasal cavity, or the tracheal tube for ventilated patients. A parallel sampling and sequencing of these anatomical parts may permit identification of bacterial contamination and of specific bacterial population in deeper samples.
Pathophysiology
Microbiome research has shown that the lungs of healthy subjects are not sterile (10), in contrast to conventional wisdom of less than a decade ago (11). The existence of distinct microbial communities within the body changes the perspective for many inflammatory and infectious diseases (10). Much research on microbial communities in disease has been performed in the gut (12). There are two important differences between the gut and the lung that result in caution when translating findings from gut microbiome research to the respiratory system. First, the bacterial load in the gut is extremely high under healthy circumstances (~108 copies/mL), while the bacterial load in the lung is extremely low as compared to the rest of body exterior (~104 copies/mL) (13). Furthermore, it is uncertain if the bacteria in a healthy lung do reproduce or are just “passing by” (11). Thus, under normal circumstances the lung has no resident microbiome that is resistant to invading microbes. Second, the gut only allows unidirectional travel by design, while all conducting airways in mammals require two-way traffic. Not only all microbes enter the lung through the mouth, must they exit that same way. Oral flora is abundant and diverse and is the healthy microbiome of the lower respiratory tract is closely related to the oral microbiome under normal circumstances (13,14).
Whiteson et al. further developed these concepts through the comparison of the oral cavity to a mainland and the lungs as island in front of the coast (15). They applied “island bibliography” to describe the diversity of the microbial community in the oral cavity and the lung. In patients with cystic fibrosis this model seemed to describe the results quite accurately (16,17). Dickson et al. further developed this idea into the “adapted island model”. In a hallmark paper, they describe three contributing factors to a change in the pulmonary microbiome: immigration, elimination and regional growth conditions (Figure 2).
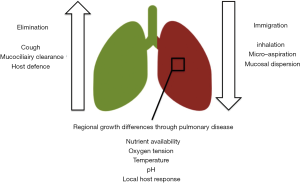
The development of VAP is generally described as an increased immigration through inhalation or micro-aspiration of pathogens into the lung. This process is facilitated through the endotracheal tube. Indeed, two studies showed a decrease in bacterial diversity in the pulmonary microbiome with prolonged mechanical ventilation (19,20). There was no association between the degree to which the diversity decreased and the development of VAP. However, in patients who developed VAP Pseudomonadales (including Pseudomonas species and Acinetobacter species) increased to a greater extent than in patients that did not develop VAP. This was irrespective of the development of colonization in the control group, which suggests that an increase in opportunistic bacteria such as Pseudomonas is associated with VAP, even if another pathogen is causing the pneumonia (19).
Elimination is also hampered in intubated and mechanically ventilated patients. Positive pressure ventilation, sedation and neuro-muscular blockage hamper the possibility to effectively cough up sputum and thereby eliminate bacteria. Positive pressure ventilation also hampers mucociliary clearance and the innate immune response is frequently disturbed in critically ill ICU-patients (21). Indeed, one study comparing patients that developed VAT to those that developed VAP found that the complement response was severely hampered in those patients progressing to pneumonia (22). This might also put the higher risk of VAP of neurosurgical patients into a perspective as they frequently have a hampered cough response and altered immune response (23).
Regional growth conditions in the lung can be the consequence of any condition that affects the lung heterogeneously. Intubation and mechanical ventilation frequently results in atelectasis of the dependent lung regions, which may put a selective pressure on specific bacteria. This might explain why a ventilation strategy with a higher positive end-expiratory pressure decreases the risk of VAP (24). Acute respiratory distress syndrome (ARDS) is characterized by protein rich pulmonary edema, which contain high level of amino acids, products of glycolysis and lipids that may form a substrate for bacterial metabolism (25-27). Empiric clinical evidence supports these experimental findings as patients with ARDS are at greater risk for VAP (28). Community-acquired pneumonia is another risk factor for VAP that may be explained through regional growth differences; the infected part of the lung is damaged and collapsed, resulting in regional conditions that may favor one pathogen. Finally, chronic pulmonary diseases such as COPD are associated with an altered respiratory microbiome in spontaneously breathing patients and it may play a role in the development of exacerbations (29-31). Alterations in the pulmonary microbiome and increased microbial loads might also be a cause for dysbiosis induced VAP; pneumonia developing by selective pressure on the existing microbiome towards the selection of a single bacterial species that continues to cause pneumonia but there is currently no evidence for this statement in ventilated patients.
Implications for prevention
Prevention of VAP mainly focuses on the prevention of immigration of bacteria into the lung. Some of the interventions that are proven to prevent VAP are; minimize sedation, supine positioning, oral hygiene measurements, maintenance of the ventilator circuit, decontamination of the mouth or gut, continuous control of cuff pressure and subglottic suctioning (32). Prevention of immigration of bacteria into the lung should, in theory, prevent colonization and VAT as well as VAP. However, these measures will not reduce the incidence of VAP in those patients in whom the lower airways do get colonized with a high number of bacteria.
Increased elimination has been tested as a prevention measure through the administration of systemic or inhaled antibiotics for the prevention of VAP. The evidence for pre-emptive treatment of patients at high risk for VAP is promising, but inconclusive until now (33-35). The available small randomized controlled trials suggest a reduction in VAP incidence with both systemic and inhaled antibiotics, but the effect on total antibiotic consumptions is unknown. A recent observational cohort study comparing selective decontamination with and without systemic antibiotic therapy suggests that a short course of cephalosporine therapy does decrease the incidence of VAP, without increasing antibiotic exposure (36). Other methods to increase elimination is through cough support or a lateral Trendelenburg position of the patient (37).
The potential of a surveillance based therapy strategy
Assuming that all pathogens that cause VAP immigrate into the lung during mechanical ventilation, they will have to pass the trachea at all times. Longitudinal studies with daily cultures of endotracheal aspirates suggest that in the majority of the patients the trachea is colonized with causative pathogen at least 1–2 days before pneumonia develops (38,39). However, colonization does frequently not progress to pneumonia. A treatment approach based on previous surveillance culture results was proposed, but was outperformed by a empirical, guideline based strategy (40). This was mainly attributed to the fact that only the results of cultures taken 3–7 days before the start antibiotic therapy were available at the time of treatment decision (40). Culture-independent detection of bacteria in endotracheal aspirates might provide faster results and thus might revive the possibility of surveillance based antibiotic therapy. The results of 16S based phylogenetic sequencing do not compare well to the results of cultures in sputum (19,20) and are therefore not useful for this purpose. More targeted assays might allow for a quantitative analysis of the number of gene copies per micro-organism (41). The major challenge with assessment of genetic material alone is that this does not indicate if it is a reproducing organism or not. Therefore, a combination of genetics with a metabolic product of the bacteria or the host immune response might be more suitable. Indeed, there is evidence suggesting that cytokines in bronchoalveolar lavage fluid or bacterial metabolites in exhaled breath can be used to diagnose VAP (42-45). The shift from culture based surveillance to a molecular assay indicative for the development of VAP could also help in earlier treatment, which might allow for a shorter course of systemic antibiotics or even the use of inhaled antibiotics alone. If the diagnostic test were to be sufficient sensitive and specific, this would save the use of longer courses of empiric, broad-spectrum antibiotics in patients suspected of VAP.
Conclusions
Microbiome research has been a hot topic in translational medicine over the past decade. Slowly, microbiome research has also been introduced to the intensive care setting. One of the areas where it may influence our pathophysiological considerations is in VAP. Many hypotheses can be made based on the adapted island model and some of these have already been tested. There is a strong need for more in-depth analyses of the changes in the microbial composition of the pulmonary microbiome during mechanical ventilation and with the development of VAP.
Acknowledgements
LD Bos is supported by a personal grant from the Dutch Lung Foundation and the presented was also influenced by a short-term fellowship supported by the European Respiratory Society.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Martin-Loeches I, Povoa P, Rodriguez A, et al. Incidence and prognosis of ventilator-associated tracheobronchitis (TAVeM): a multicentre, prospective, observational study. Lancet Respir Med 2015;3:859-68. [Crossref] [PubMed]
- Bekaert M, Timsit JF, Vansteelandt S, et al. Attributable mortality of ventilator-associated pneumonia: a reappraisal using causal analysis. Am J Respir Crit Care Med 2011;184:1133-9. [Crossref] [PubMed]
- Nair GB, Niederman MS. Ventilator-associated pneumonia: present understanding and ongoing debates. Intensive Care Med 2015;41:34-48. [Crossref] [PubMed]
- Branch-Elliman W, Wright SB, Howell MD. Determining the ideal strategy for ventilator-associated pneumonia prevention: Cost-benefit analysis. Am J Respir Crit Care Med 2015;192:57-63. [Crossref] [PubMed]
- Morgan XC, Huttenhower C. Chapter 12: Human Microbiome Analysis. PLoS Comput Biol 2012;8:e1002808. [Crossref] [PubMed]
- Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004;17:840-62. [Crossref] [PubMed]
- Scholz CF, Poulsen K, Kilian M. Novel molecular method for identification of Streptococcus pneumoniae applicable to clinical microbiology and 16S rRNA sequence-based microbiome studies. J Clin Microbiol 2012;50:1968-73. [Crossref] [PubMed]
- Goodrich JK, Di Rienzi SC, Poole AC, et al. Conducting a microbiome study. Cell 2014;158:250-62. [Crossref] [PubMed]
- Gloor GB, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol 2016;62:692-703. [Crossref] [PubMed]
- Segal LN, Rom WN, Weiden MD. Lung microbiome for clinicians: New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc 2014;11:108-16. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Martinez FJ, et al. The Microbiome and the Respiratory Tract. Annu Rev Physiol 2016;78:481-504. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011;184:957-63. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Freeman CM, et al. Bacterial Topograhy of the Healthy Human Lower Respiratory Tract. MBio 2017;8:1-12. [Crossref] [PubMed]
- Whiteson KL, Bailey B, Bergkessel M, et al. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis. Parallels from island biogeography. Am J Respir Crit Care Med 2014;189:1309-15. [Crossref] [PubMed]
- Lim YW, Schmieder R, Haynes M, et al. Mechanistic model of Rothia mucilaginosa adaptation toward persistence in the CF lung, based on a genome reconstructed from metagenomic data. PLoS One 2013;8:e64285. [Crossref] [PubMed]
- Quinn RA, Whiteson K, Lim YW, et al. Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2016;2:4. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med 2014;2:238-46. [Crossref] [PubMed]
- Zakharkina T, Martin-Loeches I, Matamoros S, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax 2017;72:803-10. [Crossref] [PubMed]
- Kelly BJ, Imai I, Bittinger K, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome 2016;4:7. [Crossref] [PubMed]
- Nakagawa NK, Franchini ML, Hila P. Mucociliary Clearance Is Impaired in Acutely Ill Patients. Chest 2005;128:2772-7. [Crossref] [PubMed]
- Martin-Loeches I, Papiol E, Almansa R, et al. Intubated patients developing tracheobronchitis or pneumonia have distinctive complement system gene expression signatures in the pre-infection period: A pilot study. Med Intensiva 2012;36:257-63. [Crossref] [PubMed]
- Hazeldine J, Lord JM, Belli A. Traumatic Brain Injury and Peripheral Immune Suppression: Primer and Prospectus. Front Neurol 2015;6:235. [Crossref] [PubMed]
- Manzano F, Fernández-Mondéjar E, Colmenero M, et al. Positive-end expiratory pressure reduces incidence of ventilator-associated pneumonia in nonhypoxemic patients. Crit Care Med 2008;36:2225-31. [Crossref] [PubMed]
- Rogers AJ, Matthay MA. Applying metabolomics to uncover novel biology in ARDS. Am J Physiol Lung Cell Mol Physiol 2014;306:L957-61. [Crossref] [PubMed]
- Evans CR, Karnovsky A, Kovach MA, et al. Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J Proteome Res 2014;13:640-9. [Crossref] [PubMed]
- Serkova NJ, Van Rheen Z, Tobias M, et al. Utility of magnetic resonance imaging and nuclear magnetic resonance-based metabolomics for quantification of inflammatory lung injury. Am J Physiol Lung Cell Mol Physiol 2008;295:L152-61. [Crossref] [PubMed]
- Forel JM, Voillet F, Pulina D, et al. Ventilator-associated pneumonia and ICU mortality in severe ARDS patients ventilated according to a lung-protective strategy. Crit Care 2012;16:R65. [Crossref] [PubMed]
- Millares L, Pérez-Brocal V, Ferrari R, et al. Functional Metagenomics of the Bronchial Microbiome in COPD. PLoS One 2015;10:e0144448. [Crossref] [PubMed]
- Segal LN, Clemente JC, Wu BG, et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 2017;72:13-22. [Crossref] [PubMed]
- Wang Z, Bafadhel M, Haldar K, et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J 2016;47:1082-92. [Crossref] [PubMed]
- Klompas M, Branson R, Eichenwald EC, et al. Strategies to prevent ventilator-associated pneumonia in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol 2014;35 Suppl 2:S133-54. [Crossref] [PubMed]
- Nseir S, Favory R, Jozefowicz E, et al. Antimicrobial treatment for ventilator-associated tracheobronchitis: a randomized, controlled, multicenter study. Crit Care 2008;12:R62. [Crossref] [PubMed]
- Poulakou G, Siakallis G, Tsiodras S, et al. Nebulized antibiotics in mechanically ventilated patients: roadmap and challenges. Expert Rev Anti Infect Ther 2017;15:211-29. [Crossref] [PubMed]
- Makris D, Luna C, Nseir S. Ten ineffective interventions to prevent ventilator-associated pneumonia. Intensive Care Med 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Bos LD, Stips C, Schouten LR, et al. Selective decontamination of the digestive tract halves the prevalence of ventilator-associated pneumonia compared to selective oral decontamination. Intensive Care Med 2017;43:1535-7. [Crossref] [PubMed]
- Li Bassi G, Torres A. Ventilator-associated pneumonia: role of positioning. Curr Opin Crit Care 2011;17:57-63. [Crossref] [PubMed]
- Ewig S, Torres A, El-Ebiary M, et al. Bacterial colonization patterns in mechanically ventilated patients with traumatic and medical head injury. Incidence, risk factors, and association with ventilator-associated pneumonia. Am J Respir Crit Care Med 1999;159:188-98. [Crossref] [PubMed]
- Berdal JE, Bjornholt J, Blomfeldt A, et al. Patterns and dynamics of airway colonisation in mechanically-ventilated patients. Clin Microbiol Infect 2007;13:476-80. [Crossref] [PubMed]
- Luna CM, Sarquis S, Niederman MS, et al. Is a strategy based on routine endotracheal cultures the best way to prescribe antibiotics in ventilator-associated pneumonia ? Chest 2013;144:63-71. [Crossref] [PubMed]
- Vincent JL, Brealey D, Libert N, et al. Rapid Diagnosis of Infection in the Critically Ill, a Multicenter Study of Molecular Detection in Bloodstream Infections, Pneumonia, and Sterile Site Infections. Crit Care Med 2015;43:2283-91. [Crossref] [PubMed]
- Conway Morris A, Kefala K, Wilkinson TS, et al. Diagnostic importance of pulmonary interleukin-1beta and interleukin-8 in ventilator-associated pneumonia. Thorax 2010;65:201-7. [Crossref] [PubMed]
- Hellyer TP, Morris AC, McAuley DF, et al. Diagnostic accuracy of pulmonary host inflammatory mediators in the exclusion of ventilator-acquired pneumonia. Thorax 2015;70:41-7. [Crossref] [PubMed]
- Fowler SJ, Basanta-Sanchez M, Xu Y, et al. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: a case-control study. Thorax 2015;70:320-5. [Crossref] [PubMed]
- Schnabel R, Fijten R, Smolinska A, et al. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci Rep 2015;5:17179. [Crossref] [PubMed]