Detection of MET amplification in gastroesophageal tumor specimens using IQFISH
Introduction
The gene mesenchymal epithelial transition factor (MET) is located on chromosome 7, which is a proto-oncogene that encodes a transmembrane receptor with intrinsic tyrosine kinase activity known as Met (or cellular-MET, c-Met). This receptor is also called the hepatocyte growth factor receptor (HGFR) after its ligand; hepatocyte growth factor (HGF). Under normal circumstances HGF activation of Met is tightly controlled by mechanisms such as paracrine ligand delivery and ligand activated receptor internalization and degradation. Despite these controls, the HGF/Met signaling contributes to oncogenesis and tumor progression in several human cancers, including gastroesophageal, colorectal, lung, breast, renal and more (1,2).
In gastroesophageal cancer, MET amplification has been reported with a prevalence of 1.5% to 30.5% (2-8), depending on the method used for the detection and the assay cut-off. MET amplification has been shown to be a factor of poor prognosis in gastroesophageal cancer (4,6,8,9). It has been suggested that MET amplification is associated with higher grade and higher stage tumors, and with a shorter median overall survival (3). The association between MET amplification and a poor prognosis in gastroesophageal cancer has also been confirmed in a recent meta-analysis (4,10). Beside the prognostics characteristics it has been suggested that MET amplification in gastroesophageal cancer potentially possess predictive properties in relation to Met targeted therapy and, thereby, could act as a companion diagnostic for some of the new therapies under development (3,11). Furthermore, in non-small cell lung cancer it has been suggested that MET amplification is involved with acquired resistance to EGFR tyrosine kinase inhibitors (12,13).
Detection of MET amplification can be performed by several techniques such as Southern blot, polymerase chain reaction (PCR), and fluorescence in situ hybridization (FISH). Here we report data on MET amplification in tumor specimens from patients with gastric (G), gastroesophageal junction (GEJ) and esophageal (E) adenocarcinomas with a MET FISH assay using the formamide-free fast IQFISH hybridization buffer (14). The purpose of this study was to investigate the inter-observer reproducibility, and to evaluate the MET signal distribution and the prevalence of MET amplification in tissue specimens from patients with G/GEJ/E adenocarcinomas.
Methods
Specimens
The study included 159 formalin-fixed and paraffin-embedded G/GEJ/E adenocarcinoma specimens. These specimens were collected consecutively from the screen population related to a phase II study with the investigational Met tyrosine kinase inhibitor AMG337 (Amgen, Thousand Oaks, CA, USA) (11). All specimens were anonymized with respect to the identity of the patients and no data on patient demographic was available. The study was conducted according to the Helsinki Declaration, and informed consent was obtained from the patients before testing was conducted. Before study initiation the protocol was reviewed and approved by the Chesapeake Institutional Review Board, Columbia, MD, USA. Evaluation of the stained slides was performed independently by two different technologists and subsequently reviewed independently by two pathologists.
MET IQFISH testing
The MET FISH staining was performed using the MET/CEN-7 IQFISH Probe Mix [for investigational use only (IUO), Agilent Technologies, Glostrup, Denmark] in combination with the Histology FISH Accessory Kit (Agilent Technologies) reagents and IQFISH staining procedure. Briefly, the FFPE specimens mounted on positively charged glass slides were exposed to heat pre-treatment using microwave oven followed by pepsin digestion at 37 °C to prepare the tissue for probe hybridization. Co-denaturation of target DNA in the specimens and the probe was done for 10 min at 66 °C followed by hybridization at 45 °C for 90 min using a Hybridizer (Agilent Technologies). The hybridization was performed simultaneously for the Texas Red-labeled DNA probe (MET) and the fluorescein-labeled PNA probe (CEN-7). The tissue specimens were subjected to stringent wash at 63 °C for 10 min before dehydration and drying. The dried slides were subsequently mounted using Fluorescence Mounting Medium containing DAPI.
The MET FISH stained slides were evaluated using a fluorescence microscope with 20× and 40× objectives. The enumeration was performed with a 100× objective. The MET/CEN-7 ratio was calculated by enumeration of 20 nuclei from the invasive tumor area. Based on the ratio, specimens were categorized into amplified (MET/CEN-7 ≥2.0) or non-amplified (MET/CEN-7 <2.0) categories. Specimens with a ratio between 1.8 and 2.2 (borderline cases) were subjected to enumeration of additional 20 nuclei and the ratio was then recalculated for the 40 nuclei to determine if amplification was present or not. Normal cells within the specimen served as an internal control for successful staining. Normal cells should exhibit the ratio expected for normal diploid cells with a one-to-one relationship of MET gene and centromere-7 signals.
MET signal distribution
Beside the evaluation of MET FISH status (amplified/non-amplified) the MET signal distribution was also evaluated. In the gastroesophageal cancer specimens, the MET signal can be classified as having a homogeneous or heterogeneous distribution. A homogeneous signal distribution is when the majority of the tumor cells in a specimen are equally amplified or equally non-amplified. For the heterogeneous signal distribution, different levels of amplification are observed in the cells in the specimen. Tumor specimens with a heterogeneous signal distribution can be sub-divided into two subcategories. If the MET amplified tumor cells are grouped together, the signal distribution is categorized as focal, whereas, if the MET amplified tumor cells in the specimen are interspersed with cells with a lower or normal MET/CEN-7 ratio, the signal distribution is categorized as mosaic. Figure 1 shows examples of the different type of signal distributions. A similar phenotypical signal distribution pattern has been described for HER2, another gene known to be amplified in a subset of patients with gastroesophageal cancer (15).

Statistical analyses
Mainly descriptive statistical methods were used. For the inter-observer reproducibility, the coefficient of variation (CV) was calculated after Log transformation of data. The Wilcoxon ranks sum test was used to test the difference in median MET/CEN-7 ratios between tumor specimens with a homogeneous and a heterogeneous signal distribution. Excel and SAS JMP PRO v11.0.0 was used for the statistical analyses.
Results
MET status and inter-observer reproducibility
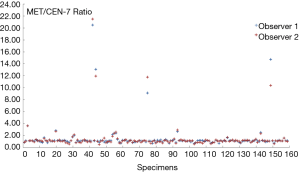
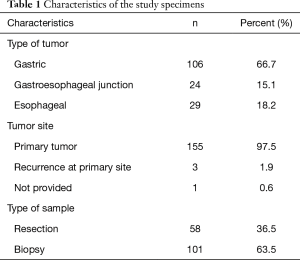
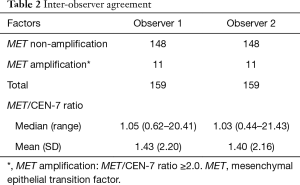
A total of 159 consecutive G/GEJ/E adenocarcinoma specimens were included in the study. An overview of the characteristics of the specimens with respect to type of tumor, sites and type of sample are given in Table 1. Two thirds (66.7%) of the specimens tested originates from the stomach and the remaining one third form esophagus or the GEJ region. Most of the specimens were biopsies (63.5%) which nearly all (97.5%) where taken from the primary tumor site. Both observers found that the same 11 out of 159 samples were MET amplified giving a prevalence of 6.9% and hence an inter-observer agreement of 100%, as shown in Table 2. Using Log data transformation, the inter-observer CV for the MET/CEN-7 ratio was estimated at 11.8% (95% CI: 10.2–13.4). The individual MET/CEN-7 ratios obtained by the two observers for the 159 tumor specimens are plotted in Figure 2. Observers agreed on signal distribution in 98.7% of specimens (157 out of 159), i.e., for one specimen observer disagreed on focal vs. mosaic heterogeneous signal distribution and for one specimen homogeneous vs. heterogeneous mosaic signal distribution.
Full table
Full table
Signal distribution
Beside the calculation of the MET/CEN-7 ratios and the evaluation of the amplification status, a qualitatively assessment of the MET signal distribution pattern was performed. This assessment showed that 12 out of the 159 tumor specimens exhibit a heterogeneous signal distribution, either focal or mosaic. Table 3 gives the prevalence of MET amplification as well as the median and the mean MET/CEN-7 ratios for the specimens according to their signal distribution. Eight out of the 12 (66.7%) specimens that had a heterogeneous signal distribution were amplified, whereas this only applied to 3 out of 147 (2.0%) for the specimens with a homogeneous signal distribution. Both the mean and the median MET/CEN-7 ratios were markedly higher for the specimens with a heterogeneous signal distribution. Comparing the median MET/CEN-7 ratios for the two groups showed a statistically significantly higher value for the specimens with a heterogeneous signal distribution compared to the specimens with a homogeneous signal distribution (P<0.0001, Wilcoxon rank sum test).

Full table
Discussion
In the current study, MET amplification was detected in 6.9% of the 159 consecutive specimens originating from individual patients with gastroesophageal cancer. The vast majority of the tested specimens were primary tumors and mainly taken as biopsies from the stomach. Comparing the prevalence of MET amplification in this study with the relatively few data available in literature, the prevalence seems to be in the lower end of the reported range. For gastroesophageal cancer, the prevalence of MET amplification has been reported in the range of 1.5% to 30.5% (2-8). However, when such data is compared, it is important also to take the analysis method used and the assay cut-off into consideration. The highest prevalence reported originate from Southern blot analyses and PCR based assays as these methods are unable to distinguish between true gene amplification and polysomy, which then result in higher estimates for the prevalence of MET gene amplification (5). The MET amplification prevalence of 6.9% observed in this study is within the range 1.5% to 8.3% found in other studies by means of FISH (2,5,6).
The overall inter-observer agreement with respect to the MET status (amplified/non-amplified) was 100%. The two observers detected the same 11 tissues specimens as being MET amplified. Furthermore, the median and mean MET/CEN-7 ratios obtained by the two observers were almost identical as can be seen from Table 2. The largest differences between the two observers were found for the specimens that had the highest MET/CEN-7 ratios, as shown in Figure 2, where enumeration of the MET hybridization signals could be challenging. However, these differences have no influence on the MET status determination, with regards to amplified or non-amplified, as they lie far above the assay cut-off at a MET/CEN-7 ratio of 2.0. Discrepancy between observers for the higher gene/centromere ratios has also been reported for HER2 using the FISH method (16,17), and is due to the nature of enumerating signals in the nuclei. When high amplification levels are present, signals may lie in clusters, where it is difficult or impossible to enumerate the precise signal numbers. In such situations, a qualified estimate of every cluster in the relevant nuclei is performed, which will introduce variation between observers. The inter-observer CV was estimated at 11.8%, which is in the lower end of previous reported CVs for this type of assay (17).
As part of the study a qualitatively assessment of the MET signal distribution pattern at the tissue level was performed. When the 159 tumor specimens were divided into homogeneous and heterogeneous signal distribution patterns, as shown in Table 3, the prevalence of MET amplification was much higher in the group of patients who harbored tumors with a heterogeneous signal distribution compared to the group with a homogeneous signal distribution. In the group with a heterogeneous signal distribution 8 out of the 12 (66.7%) tumor specimens were amplified, whereas this only was the situation for 3 out of 147 (2.0%) for the tumor specimens with a homogeneous signal distribution. We also found that the median MET/CEN-7 ratio for the tumor specimens with a heterogeneous signal distribution was significantly higher than for the tumor specimens with a homogeneous signal distribution.
Intra tumor heterogeneity and tumor aneuploidy are important mechanisms for development of drug resistance, which possess major challenges in relation to anti-cancer therapy (18). The different in situ hybridization techniques offers the possibility of combining genetic/chromosomal information with spatial phenotypic characteristics of the tumor. The FISH data from the current study indicate that a specific phenotypic characteristic is linked to MET amplification, which could suggest that the MET positive tumor cells are dominated by a heterogeneous signal distribution pattern in gastroesophageal cancer. Recent published data on the HER2 gene has likewise shown a similar link between heterogeneous signal distribution and gene amplification and hence the associated tumor growth pattern (15).
To the best of our knowledge this relationship between MET amplification and a heterogeneous signal distribution has not previously been described for gastroesophageal cancer. As this specific relationship, also has been shown for HER2 this might be a common feature linked to gene amplification in gastroesophageal cancer (15). However, further studies will be needed to confirm these findings and show if this relationship will have prognostic and/or predictive implications for patients with MET amplified gastroesophageal cancer. Furthermore, we underline that the hypothesis raised about the link between MET amplification and the heterogeneous signal distribution in gastroesophageal cancer is generated on a relatively small number of tumor specimens and therefore must be validated in a larger study.
Conclusions
The novel FISH assay showed a high inter-observer reproducibility both with respect to MET status and the signal distribution pattern. Based on the finding in the study it is suggested that MET amplification mainly is associated with tumor cells that is represented by a heterogonous growth pattern. However, due to the relative small sample size in the study the signal distribution pattern findings require further validation.
Acknowledgements
None.
Footnote
Conflicts of Interest: N Go is an employee of Amgen. KB Nielsen, J Mollerup, and A Jepsen are employees of Agilent Technologies. JT Jørgensen has worked as a consultant for Agilent Technologies, Euro Diagnostica, and Oncology Ventures and has given lectures at meetings sponsored by AstraZeneca, Merck Sharp & Dohme, and Roche.
Ethical Statement: The study protocol was approved by the Chesapeake Institutional Review Board (No. Pro00009139), approval date December 19, 2013. Written informed consent was obtained from the patients before testing was conducted.
References
- Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signaling pathway in cancer therapy. Expert Opin Ther Targets 2012;16:553-72. [Crossref] [PubMed]
- Jardim DL, Tang C, Gagliato Dde M, et al. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I Clinic. Clin Cancer Res 2014;20:6336-45. [Crossref] [PubMed]
- Lennerz JK, Kwak EL, Ackerman A, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 2011;29:4803-10. [Crossref] [PubMed]
- Peng Z, Zhu Y, Wang Q, et al. Prognostic significance of MET amplification and expression in gastric cancer: a systematic review with meta-analysis. PLoS One 2014;9:e84502. [Crossref] [PubMed]
- Kawakami H, Okamoto I, Okamoto W, et al. Targeting MET Amplification as a New Oncogenic Driver. Cancers (Basel) 2014;6:1540-52. [Crossref] [PubMed]
- An X, Wang F, Shao Q, et al. MET amplification is not rare and predicts unfavorable clinical outcomes in patients with recurrent/metastatic gastric cancer after chemotherapy. Cancer 2014;120:675-82. [Crossref] [PubMed]
- Ooi A, Oyama T, Nakamura R, et al. Semi-comprehensive analysis of gene amplification in gastric cancers using multiplex ligation-dependent probe amplification and fluorescence in situ hybridization. Mod Pathol 2015;28:861-71. [Crossref] [PubMed]
- Matsusaka S, Kobunai T, Yamamoto N, et al. Prognostic impact of KRAS mutant type and MET amplification in metastatic and recurrent gastric cancer patients treated with first-line S-1 plus cisplatin chemotherapy. Genes Cancer 2016;7:27-35. [PubMed]
- Graziano F, Galluccio N, Lorenzini P, et al. Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol 2011;29:4789-95. [Crossref] [PubMed]
- Erichsen R, Kelsh MA, Oliner KS, et al. Prognostic impact of tumor MET expression among patients with stage IV gastric cancer: a Danish cohort study. Ann Epidemiol 2016;26:500-3. [Crossref] [PubMed]
- Hughes PE, Rex K, Caenepeel S, et al. In Vitro and In Vivo Activity of AMG 337, a Potent and Selective MET Kinase Inhibitor, in MET-Dependent Cancer Models. Mol Cancer Ther 2016;15:1568-79. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009;20:298-304. [Crossref] [PubMed]
- Matthiesen SH, Hansen CM. Fast and non-toxic in situ hybridization without blocking of repetitive sequences. PLoS One 2012;7:e40675. [Crossref] [PubMed]
- Jørgensen JT, Nielsen KB, Kjærsgaard G, et al. Gene Signal Distribution and HER2 Amplification in Gastroesophageal Cancer. J Cancer 2017;8:1517-24. [Crossref] [PubMed]
- Umemura S, Osamura RY, Akiyama F, et al. What causes discrepancies in HER2 testing for breast cancer? A Japanese ring study in conjunction with the global standard. Am J Clin Pathol 2008;130:883-91. [Crossref] [PubMed]
- Viale G, Paterson J, Bloch M, et al. Analysis of HER2 status in gastroesophageal tumor specimens using a new automated HER2 IQFISH pharmDx™ (Dako Omnis) assay. Histol Histopathol 2016;31:1327-35. [PubMed]
- McGranahan N, Burrell RA, Endesfelder D, et al. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 2012;13:528-38. [Crossref] [PubMed]