Meta-analysis of the association between CD33 and Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) is a complicated neurodegenerative disease with progressive cognitive impairment common in elderly people. In 2006, the prevalence of AD was 26.6 million around the world, and the number of AD will quadruple by 2050 (1). AD consists of early-onset AD (EOAD) and late-onset AD (LOAD). LOAD, which accounts for the majority of AD, is the result of interaction between environmental and genetic factors (2). Genetic factors play an important role in it, and the heritability is estimated to be up to 80% (3-6). Up to now, the apolipoprotein E (APOE) gene is the only one gene that was certainly recognized to increase the risk of LOAD, but the APOE ε4 gene can only interpret 27.3% about the risk of AD onset (7-10). Therefore, further efforts are needed to look for risk genes other than APOE.
The cluster of differentiation 33 (CD33) gene is an immune function protein located in 19p13.33 with several functions, such as cell adhesion, anti-inflammatory signaling, and endocytosis functions (11). In addition, it is one of the members of the sialic acid-binding Ig-like lectin (SIGLEC) family (12). Several studies have investigated the important role of CD33 rs3865444 polymorphism in AD. As for rs3865444, the risk allele (C) was related with the overexpression of CD33 on mononuclear cell surface, which is involved in the pathogenesis of AD through down-regulation of β-amyloid (Aβ) internalization, accumulation of neuritic amyloid pathology, and regulation of microglia levels (13). Both lines of evidence indicate that CD33 gene could play an important role in susceptibility to AD.
In 2011, three large-scale genome-wide association studies (GWAS) conducted by Hollingworth et al. (14), Carrasquillo et al. (15), and Naj et al. (16) confirmed that the gene CD33 showed significantly correlation with LOAD in Europe. After that, the associations between AD and the variants of CD33 have become the focus of many studies. But the conclusions of these studies differ from each other. But the conclusions of these studies are inconsistent (7,14-29). To better illuminate the associations between CD33 and the susceptibility to AD, a meta-analysis was conducted by analyzing and summarizing the relevant studies.
Methods
Literature search
We searched literature in PubMed, EMbase, and Cochrane library up to 1 October 2017. Medical Subject Heading (MESH) terms were used: (CD33) AND {[(Alzheimer’s) OR dementia] OR Alzheimer disease}. Additional studies were screened manually in each qualifying study.
Inclusion criteria
We chose the studies meeting the following criteria: (I) studies must evaluate the associations between polymorphism of CD33 and the susceptibility to AD; (II) the research type of the selected studies was case-control design; (III) adequate information should be accessible, such as the study sample size of each research group, allele or genotype frequencies, OR, 95% CI and the P value; (IV) the diagnosis of AD should meet the clinical criteria set by the World Health Organization.
Data extraction
According to the specified selection criteria, all data were extracted independently by two investigators. The following data were extracted: (I) name of the first author, (II) publication year, (III) country and ethnicity of origin, (IV) numbers of cases and controls, (V) allele frequency distributions and numbers of different genotypes, (VI) the OR with 95% CI of CD33. Newcastle-Ottawa quality scale (NOS) was selected to evaluate the quality of eligible studies: (I) the selection; (II) the comparability; (III) the exposure. Studies with a score larger than seven points were considered to be of high quality.
Statistical analysis
Stata 11.0 software version 11.0 (StataCorp LP, College Station, TX, USA) and R software [Version 3.2.2, Copyright(C) 2014 The R Foundation for Statistical Computing] were selected for statistical analysis. Hardy-Weinberg equilibrium (HWE) was used to verify the representation of the selected studies. We conducted a subgroup analyses on the basis of ethnicity (Asian, Caucasian and Negroid). Statistical heterogeneity could be evaluated by Cochran’s Q statistic and I2 Statistic test. When significant heterogeneity (PQ >0.10 and I2>50%) was observed, the random-effects model was carry out; in addition, the fixed-effects model was adopted. In order to assess the stability of the results, we conducted sensitivity analysis by omitting a single study each time. Funnel plots were generated by the R software to evaluate the potential publication bias. And publication bias was also checked by Egger’s linear regression analysis and Begg’s rank correlation test, and P<0.05 indicated a significant publication bias.
Results
Study inclusion and characteristics
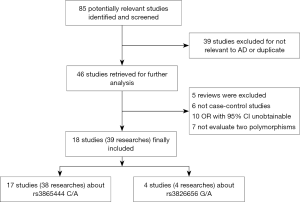
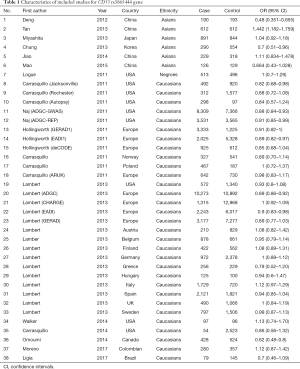
In the present meta-analysis, a systematic overview of relevant studies to evaluate the risk between gene CD33 and the susceptibility to AD have been accomplished. Eighty-five articles were searched from PubMed, EMbase, Cochrane library and other sources, such as using a manual retrieval from relevant studies. After the initial scan of titles and abstracts, 39 unrelated studies were excluded because they were not relevant to AD or they were duplicate publications. According to the inclusion standards set in advance, 28 publications were excluded. After our screening, 18 articles on relevant SNPs were included in this meta-analysis (Figure 1). Eventually, 38 researches investigating the single nucleotide polymorphism (SNP) of rs3865444 and 4 researches investigating the SNP of rs3826656 were included. The characteristics of these involved studies were presented in the Tables 1,2.
Full table

Full table
CD33 rs3865444 SNP and AD susceptibility
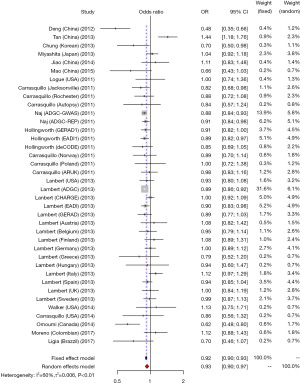
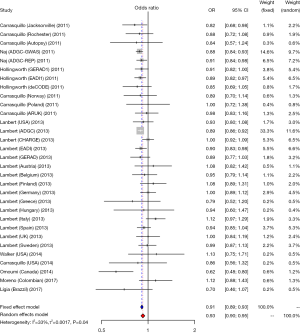
We studied the correlation between SNP rs3865444 of CD33 and AD susceptibility in this meta-analysis. Among the 38 enrolled researches, 31 were performed in Caucasian population, 6 were performed in Asian population and only one was performed in Negroid population. In the pooled OR, we can observe significant heterogeneity. We used a random-effect model to assess the overall effect of the C/A allele model (I2=60%, P<0.01), and the A mutant of rs3865444 was found to be significantly associated with AD prevention (OR =0.93; 95% CI, 0.90–0.97; Figure 2). Considering the difference of ethnicity, we made subgroup analyses. The existence of high heterogeneity could be identified. In the stratified analysis among Caucasian ethnicity, the heterogeneity was decreased (I2=33%, P=0.04), and a fixed-effect model could be used. Similarly, the association of rs3865444 and AD susceptibility was not verified (OR =0.91; 95% CI, 0.89–0.93; Figure 3). Besides, in the stratified analysis among Asian ethnicity, the heterogeneity was increased (I2=88%, P<0.01), and a random-effect model may be used. But the association between rs3865444 and AD susceptibility turned into no significance (OR =0.87; 95% CI, 0.65–1.17; Figure 4).
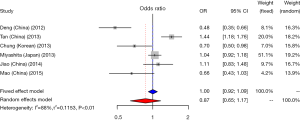
CD33 rs3826656 SNP and AD susceptibility
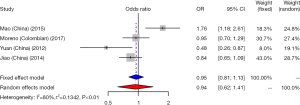
We did a research about the association between CD33 rs3826656 SNP and AD susceptibility in a cumulative meta-analysis. Among the 4 studies on rs3826656, 3 were conducted in the Asian population and only one was performed in Caucasians. We used a random-effect model to evaluate the effect of the G/A allele model (I2=80%, P<0.01). The pooled ORs identified no correlation between SNP rs3826656 and AD in the present meta-analysis (OR =0.94; 95% CI, 0.62–1.41; Figure 5).
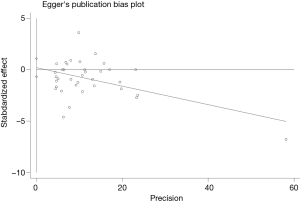
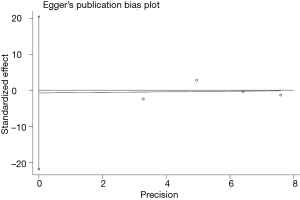
Publication bias and sensitivity analysis
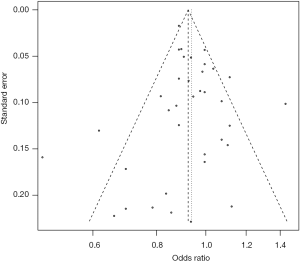
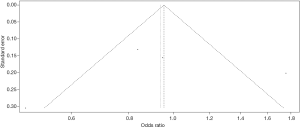
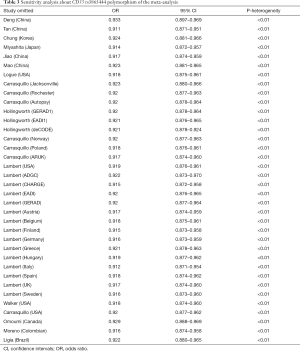
The funnel plot for the two SNPs of CD33 exhibited relative symmetry (Figures S1,S2). There were no statistically significant publication bias in SNP rs3865444 (PBegg’s =0.186, PEgger’s =0.643; Figure S3). Similarly, there was no publication bias in SNP rs3826656 (PBegg’s =1, PEgger’s =0.892; Figure S4). Sensitivity analysis of CD33 rs3865444 SNP was performed by continuously excluding individual studies to test the effect of individual data on the pooled ORs, of which the results revealed that no individual study can significantly affect the overall value of ORs and 95% CIs (Table 3). As for CD33 rs3826656 SNP, the results remained as insignificant as before.
Full table
Discussion
Our meta-analysis summarized the evidence to date of the links between common polymorphisms of CD33 gene and the risk of AD. The variant of rs3865444 was identified to be significantly associated with lower risk of AD, while no correlation was showed of the rs3826656 SNP with the risk of AD. We believe that our findings containing the latest published studies will be meaningful for future genetic studies on AD and CD33 gene, which may be a potential candidate gene for AD susceptibility.
Since July 2011, related studies on the association between the CD33 gene polymorphisms and AD have been carried out all over the world with subjects in different populations, such as Caucasian, Asian and Negro populations. Because of the few number of articles about Negroid, no subgroup analysis on it was done. In China, Tan et al. in 2013 showed that the polymorphism of CD33 rs3865444 was a risk factor for AD (OR =1.442; 95% CI, 1.182–1.759) (17), which was different from results in most Caucasian population. But in China, another study conducted by Deng et al. obtained an opposite conclusion compared with that of Tan et al. (OR =0.48; 95% CI, 0.351–0.655) (20). In 2013, a meta-analysis conducted by Lambert showed the evidence that the variants of rs3865444 were found to be significantly associated with lower risk of AD in Caucasian population (OR =0.94; 95% CI, 0.91–0.96) (30). Considering the inconsistent results, ethnic subgroups were used to eliminate possible ethic differences. As for the polymorphism of CD33 rs3826656, Mao et al. made a meta-analysis in China to identify whether SNP rs3826656 was associated with increased risk of AD (OR =1.39; 95% CI, 1.09–1.76) (31). In order to further harmonize these contradictory findings and to acquire a more accurate conclusion about the two SNPs of CD33 gene, an updated meta-analysis was conducted.
Based on those recent studies on the role of rs3865444 SNP in pooled population, our results showed a significant link between risk allele (A) and lower risk of AD. In a subgroup analysis, the results of the present study in Caucasian population supported that rs3865444 variant had protective association with AD. As predicted, the result was the same as that of the study conducted by Lambert (30). In Asian population, the correlation between rs3865444 gene polymorphism and the incidence of AD could not be determined. We couldn’t agree with the conclusions made by Tan et al. and Deng et al. Besides, there was only one study about the population of Negroid ethnicity with limitation of the sample size, which might not provide sufficient data in Negroid population. Large-scale case-control studies in different ethnicities and regions need to be performed. With regard to rs3826656 gene polymorphism, there was no association between this gene locus and AD, which was different from the result drawn by Mao et al. (31). To our knowledge, our study offers a more systematic and higher-quality evidence to indicate the relationship between two SNPs of CD33 and AD. In addition, we added several new studies which increased the total amount of subjects to raise the accuracy of outcomes. All included studies were published in authoritative journals after 2011, which identified that the research on the CD33 gene polymorphisms were promising and novel.
We have to admit that some limitations in our meta-analysis should be illuminated. There were lots of polymorphisms in CD33, such as rs10419982 (32), rs273652 (22), rs35112940 (33) and so on. Because of lack of researches on these loci, our study, which only concentrated on two polymorphisms of CD33, may not fully represent the function of CD33 gene. A larger range of studies are demanded to reinforce the representation of this gene.
It is likely that the CD33 polymorphism will play an important role on the way to genetic susceptibility in the future. The effective therapeutic interventions for AD are still very limited and, therefore, we hope further research on gene CD33 can offer a specific structural goal for the development of novel therapeutic approaches.
Conclusions
The SNP rs3865444 of CD33 played a protective role in AD, and this protective role still existed in Caucasian population in the subgroup analysis. There were no association between SNP rs3826656 and AD. Further studies with large sample size should be done to confirm our findings.
Acknowledgements
This work was supported by grants from Qingdao Key Health Discipline Development Fund, Qingdao Outstanding Health Professional Development Fund, and Shandong Provincial Collaborative Innovation Center for Neurodegenerative Disorders.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Brookmeyer R, Johnson E, Ziegler-Graham K, et al. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement 2007;3:186-91. [Crossref] [PubMed]
- Lv ZY, Tan CC, Yu JT, et al. Spreading of Pathology in Alzheimer's Disease. Neurotox Res 2017;32:707-22. [Crossref] [PubMed]
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006;63:168-74. [Crossref] [PubMed]
- Palotas A, Kalman J. Candidate susceptibility genes in Alzheimer's disease are at high risk for being forgotten-- they don't give peace of mind. Curr Drug Metab 2006;7:273-93. [Crossref] [PubMed]
- Antoniades D, Katopodi T, Pappa S, et al. The role of reelin gene polymorphisms in the pathogenesis of Alzheimer's disease in a Greek population. J Biol Regul Homeost Agents 2011;25:351-8. [PubMed]
- Wang LZ, Tian Y, Yu JT, et al. Association between late-onset Alzheimer's disease and microsatellite polymorphisms in intron II of the human toll-like receptor 2 gene. Neurosci Lett 2011;489:164-7. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants in ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Jayadev S, Leverenz JB, Steinbart E, et al. Alzheimer's disease phenotypes and genotypes associated with mutations in presenilin 2. Brain 2010;133:1143-54. [Crossref] [PubMed]
- Alzheimer’s Association. 2015 Alzheimer's disease facts and figures. Alzheimers Dement 2015;11:332-84. [Crossref] [PubMed]
- Liu Y, Tan L, Wang HF, et al. Multiple Effect of APOE Genotype on Clinical and Neuroimaging Biomarkers Across Alzheimer's Disease Spectrum. Mol Neurobiol 2016;53:4539-47. [Crossref] [PubMed]
- Wang WY, Liu Y, Wang HF, et al. Impacts of CD33 Genetic Variations on the Atrophy Rates of Hippocampus and Parahippocampal Gyrus in Normal Aging and Mild Cognitive Impairment. Mol Neurobiol 2017;54:1111-8. [Crossref] [PubMed]
- Malik M, Simpson JF, Parikh I, et al. CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci 2013;33:13320-5. [Crossref] [PubMed]
- Zhu JB, Tan CC, Tan L, et al. State of Play in Alzheimer's Disease Genetics. J Alzheimers Dis 2017;58:631-59. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Carrasquillo MM, Belbin O, Hunter TA, et al. Replication of EPHA1 and CD33 associations with late-onset Alzheimer's disease: a multi-centre case-control study. Mol Neurodegener 2011;6:54. [Crossref] [PubMed]
- Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436-41. [Crossref] [PubMed]
- Tan L, Yu JT, Zhang W, et al. Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement 2013;9:546-53. [Crossref] [PubMed]
- Dos Santos LR, Pimassoni LHS, Sena GGS, et al. Validating GWAS Variants from Microglial Genes Implicated in Alzheimer's Disease. J Mol Neurosci 2017;62:215-21. [Crossref] [PubMed]
- Omoumi A, Fok A, Greenwood T, et al. Evaluation of late-onset Alzheimer disease genetic susceptibility risks in a Canadian population. Neurobiol Aging 2014;35:936.e5-12. [Crossref] [PubMed]
- Deng YL, Liu LH, Wang Y, et al. The prevalence of CD33 and MS4A6A variant in Chinese Han population with Alzheimer's disease. Hum Genet 2012;131:1245-9. [Crossref] [PubMed]
- Reitz C, Jun G, Naj A, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4, and the risk of late-onset Alzheimer disease in African Americans. JAMA 2013;309:1483-92. [Crossref] [PubMed]
- Chung SJ, Lee JH, Kim SY, et al. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis Assoc Disord 2013;27:250-7. [Crossref] [PubMed]
- Zhang DF, Li J, Wu H, et al. CFH Variants Affect Structural and Functional Brain Changes and Genetic Risk of Alzheimer's Disease. Neuropsychopharmacology 2016;41:1034-45.
- Yuan Q, Chu C, Jia J. Association studies of 19 candidate SNPs with sporadic Alzheimer's disease in the North Chinese Han population. Neurol Sci 2012;33:1021-8. [Crossref] [PubMed]
- Jiao B, Liu X, Zhou L, et al. Polygenic Analysis of Late-Onset Alzheimer’s Disease from Mainland China. PLoS One 2015;10:e0144898. [PubMed]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013;45:1452-8. [Crossref] [PubMed]
- Walker DG, Whetzel AM, Serrano G, et al. Association of CD33 polymorphism rs3865444 with Alzheimer's disease pathology and CD33 expression in human cerebral cortex. Neurobiol Aging 2015;36:571-82. [Crossref] [PubMed]
- Miyashita A, Koike A, Jun G, et al. SORL1 is genetically associated with late-onset Alzheimer's disease in Japanese, Koreans and Caucasians. PLoS One 2013;8:e58618. [Crossref] [PubMed]
- Carrasquillo MM, Khan Q, Murray ME, et al. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurology 2014;82:1455-62. [Crossref] [PubMed]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452-8. [Crossref] [PubMed]
- Mao YF, Guo ZY, Pu JL, et al. Association of CD33 and MS4A cluster variants with Alzheimer's disease in East Asian populations. Neurosci Lett 2015;609:235-9. [Crossref] [PubMed]
- Logue MW, Schu M, Vardarajan BN, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol 2011;68:1569-79. [Crossref] [PubMed]
- Sims R, van der Lee SJ, Naj AC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet 2017;49:1373-84. [Crossref] [PubMed]


