Prostate cancer in the era of “Omic” medicine: recognizing the importance of DNA damage repair pathways
Introduction
Prostate cancer remains one of the most commonly diagnosed cancers for men in the Western world and a leading cause of cancer-related deaths worldwide (1). Despite the progress in prostate cancer screening and in the management of localized disease, metastatic prostate cancer is invariably fatal. In the United States alone, the National Cancer Institute estimates that over 26,000 men will have died from prostate cancer in 2017 (2).
Since Huggins and Hodges discovered the effectiveness of hormonal therapy in treating prostate cancer more than 75 years ago, androgen deprivation therapy (ADT) has remained the backbone of systemic prostate cancer treatment. Unfortunately, many patients with hormone-sensitive prostate cancer will eventually develop progressive disease despite castrate levels of serum androgens, a disease state known as castrate-resistant prostate cancer (CRPC). In addition, the clinical risk posed by advanced prostate cancer is also determined by whether tumor cells have colonized other organs, a process known as metastasis, which can occur in both the hormone-sensitive setting, or more commonly, in CRPC.
Although metastatic CRPC is incurable, several current treatments extend overall survival. Currently approved therapies in the United States include second-generation anti-androgens (e.g., enzalutamide and abiraterone), radium-223, immunotherapy (sipuleucel-T) and chemotherapy (docetaxel, cabazitaxel and mitoxantrone) (3-8). However, each of these therapies prolongs overall survival by only a few months. In addition, unlike cancers where molecular alterations inform treatment decisions [for example, using EGFR inhibitors for certain forms of EGFR-mutated non-small cell lung cancer (9)], treatment for prostate cancer is still largely administered without consideration of the underlying genomic alterations within each individual patient’s tumor.
Recently, a number of collaborative studies have further defined the molecular landscape of advanced prostate cancer (10-12). [For the purposes of this review, we will focus on androgen receptor (AR)-positive prostate adenocarcinomas, which comprise the vast majority of prostate cancers; while emerging evidence suggests that there are several other distinct phenotypic entities including small cell, neuroendocrine prostate cancer (NEPC) as well as AR-null, non-NEPC prostate cancer, the molecular characteristics of these less common subtypes are still being defined (13-15)]. Efforts to sequence and understand the genomes, exomes, epigenomes and transcriptomes have revealed common aberrations in genes such as the AR, ETS (E26 transformation specific) gene fusions, TP53 and PTEN, as well as perturbations in pathways involving WNT/β-catenin signaling, cell cycle regulation and DNA repair. In addition, prostate cancer can be molecularly grouped into luminal and basal subtypes, which are prognostic and associated with differential response to ADT (16). Furthermore, genomic predictors can identify patients whose tumors might have enhanced responses to post-operative radiation therapy (17). Taken together, these molecular studies demonstrate that different prostate tumor subtypes harboring different molecular mutations have varying responses to current therapeutic interventions. Therefore, identifying dysregulated pathways in each individual patient’s tumor and choosing an appropriate therapy may enhance responses and improve survival. However, there are no trials to date that show a survival benefit of using prostate cancer genomics to guide treatment trajectory.
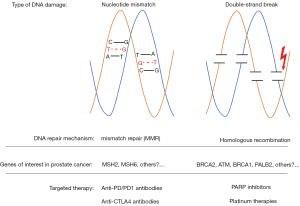
The DNA damage repair (DDR) response, integral in maintaining genomic stability and integrity, has recently emerged as an important contributor to prostate cancer pathogenesis (18). Many of the key components of the DDR are tumor suppressors, which prevent the formation and propagation of mutations and copy-number alterations. The DDR system includes multiple distinct pathways, two of which are homologous recombination (HR), which relies on BRCA1, BRCA2 and ATM, and mismatch repair (MMR), which involves MLH1, MSH2, MSH6 and PMS2 (Figure 1). When genes involved in DDR are mutated, genomic instability and increased mutational burden can occur, which can contribute to continued tumor growth. However, we are beginning to learn that these DDR mutations can also be the “Achilles’ heel”, rendering the cancer cells more sensitive to alternative approaches and targeted therapy. In this Review, we summarize our current understanding of DDR deficiencies in prostate cancer, and discuss how this is changing the treatment landscape of the disease. We highlight several ongoing studies targeting DNA repair deficiency in prostate cancer and propose future studies that may open additional opportunities for targeted approaches. Finally, we highlight the resistance mechanisms that may emerge as we begin to target these pathways individually and discuss potential combinatorial approaches that may delay or prevent resistance, as our knowledge of tumor biology continues to grow.
Homologous repair deficiencies in prostate cancer
HR is one of two pathways that eukaryotic cells use to repair DNA damage when double-stranded breaks (DSBs) occur. This process, which occurs during the synthesis, or S phase, of the cell cycle, involves using the sister chromatid as the template for DNA synthesis to repair DSBs. HR depends on multiple proteins (including BRCA1, BRCA2, ATM, RAD51, and PALB2 and others) to (I) detect DSBs, (II) resect the 3’ ends near the break, (III) promote pairing and invasion of the homologous double helix, and (IV) synthesize the corresponding DNA sequence (19). As this process relies on the sister chromatid to provide the “good” copy, HR is characterized by a low mutation rate and maintains fidelity. Alternatively, cells can utilize another pathway to repair DSBs, a process called non-homologous end joining (NHEJ). Unlike HR, NHEJ is much more error-prone, and is characterized by frequent insertions and deletions in the damaged region of the genome, leading to increased genomic instability (20,21). Deficiencies in HR are the most common DDR defects found in prostate cancer, and without a functioning HR pathway, tumor cells rely exclusively on NHEJ (10). Accordingly, Brca1 or Brca2 deficiency in mouse models leads to a mutational signature characterized by deletions followed by tandem repeats (20-22). Interestingly, BRCA1, BRCA2, and RAD51 also have HR-independent functions that help to prevent genomic instability and tumorigenesis (23).
Loss of function mutations in DDR genes (both germline and somatic) have been increasingly recognized in the progression and pathogenesis of aggressive prostate cancer. Recent studies demonstrate an increased prevalence of HR mutations in metastatic castration resistant prostate cancer (mCRPC), with approximately 20–25% of patients with metastatic disease having germline or somatic mutations in BRCA2, ATM, or BRCA1 compared to <10% found in primary prostate tumors (10,11,18,24). This enrichment in metastatic CRPC patients points towards an important role of HR deficiency in the development of advanced disease. Accordingly, familial inherited mutations in BRCA2 and BRCA1 confer an increased risk of developing aggressive prostate cancer. For example, heterozygous germline BRCA2 mutations confer an 8.6-fold increased risk of prostate cancer in men less than 65 years old, while germline BRCA1 mutations confer a 3.75-fold increased risk (25,26). In addition, men with deleterious BRCA2 germline mutations are diagnosed at a younger age, have more advanced tumor stage and grade at diagnosis, experience shorter median overall survival, and have increased risk of prostate cancer specific mortality (27). Therefore, it is important to systematically identify these patients, and refer them to genetic counselors as appropriate to discuss implications for their family members.
Targeting homologous repair defects
An encouraging development in the last decade has been the identification of novel therapeutic strategies to treat tumors with HR deficiency (28,29). Although detecting HR deficiency with enough sensitivity and specificity remains difficult, there is a clear distinct genomic phenotype that can be observed (30). Importantly, tumors deficient in HR have increased sensitivity to agents that lead to DSBs such as platinum-based chemotherapies and poly-ADP-ribose polymerase (PARP) inhibitors (31-34). Platinum-based chemotherapy such as cisplatin and carboplatin are DNA cross-linking agents that halt the progression of replication forks whereas PARP inhibitors prevent the normal release of the DNA repair protein PARP1 from DNA, leading to replication fork stalling and eventual DSBs. Without a functioning HR pathway, the accumulation of dsDNA damage renders cells non-viable.
In other cancer types, there is evidence that BRCA1 and BRCA2 mutations sensitize tumors to treatment with platinum-based chemotherapies (35-37). Several phase II combination therapy trials in prostate cancer have been performed to study the efficacy of carboplatin with docetaxel, paclitaxel, everolimus, and etoposide (38-41). These studies showed a PSA50 (i.e., reduction of PSA by 50%) response rate ranging from 18–26% in mCRPC patients. Although tumor sequencing was not performed to identify patients with DDR deficiencies in this population, this frequency is close to the observed rate of HR deficiency observed in metastatic CRPC (41-43). Although it is entirely speculative, this suggests the possibility of positive responses to platinum in HR-deficient metastatic CRPC patients (44). In particular, non-specific DNA-damaging agents such as platinum agents may prove to be a promising treatment modality for HR-deficient CRPC patients, as they are reasonably well-tolerated, inexpensive, and their efficacy does not rely solely on disrupting a single mechanism of DNA repair. Indeed, several ongoing trials are investigating platinum-based chemotherapy in prostate cancer patients with known DDR defects (Table 1).

Full table
Based on the success of PARP inhibitor therapies in breast and ovarian cancer patients with HR deficiency, recent trials have investigated the efficacy of PARP inhibitors in the context of metastatic CRPC patients who carry deleterious HR mutations. PARP is a nuclear enzyme complex that functions in repairing single-strand DNA breaks (SSBs) and coordinating DNA repair through base excision. PARP contains a zinc-finger DNA-binding domain that detects the SSBs and then catalyzes the transfer of ADP-ribose to several protein acceptors involved in DNA metabolism and base excision repair. Therefore, PARP inhibition impairs the base-excision repair (BER) pathway, resulting in the accumulation of DNA damage. The TOPARP-A trial was carried out in the United Kingdom as an open label, single-arm, phase II trial to evaluate the efficacy of 400 mg olaparib twice a day in men with advanced metastatic CRPC (24). Patients were eligible if they experienced clinical progression after one or two lines of chemotherapy (with no prior exposure to platinum, cyclophosphamide, mitoxantrone, or PARP inhibitors). The primary endpoint was a reduction of PSA by 50% or the conversion of circulating tumor-cell count from ≥5 cells/7.5 mL of blood, to <5 cells/7.5 mL of blood. These patients were heavily pre-treated—all participants had received prior docetaxel, 49 of 50 (98%) had received abiraterone or enzalutamide, and 29 (58%) had received cabazitaxel. Fourteen of 16 patients (88%) who responded to olaparib treatment were found to have a DDR gene mutation. All 7 patients with BRCA2 loss, and 4 of the 5 patients with ATM deficiency responded to olaparib. Interestingly, of the patients with BRCA2 deficiency, 4 patients had bi-allelic somatic loss, while 3 patients had deleterious germline mutations (24). The authors concluded that treatment with a PARP inhibitor in patients with defects in DNA repair leads to high response rates.
In addition to olaparib, several other PARP inhibitors are currently being tested in clinical trials for prostate cancer. For example, TRITON II and TRITON III are ongoing trials testing the PARP inhibitor rucaparib. TRITON II is a multi-center phase II study evaluating the efficacy of rucaparib monotherapy in patients with metastatic CRPC and HR deficiency who have progressed after taxane treatment and a next generation AR inhibitor. This trial includes patients harboring a deleterious mutation in one of classical DDR genes (i.e., BRCA1, BRCA2, or ATM) as well as those harboring a mutation in an expanded HR deficiency panel (including BRIP1, BARD1, CHEK2, PALB2, NBN and CDK12). Intriguingly, these non-BRCA1/2 genes may account for up to 35% of inherited DDR mutations in metastatic CRPC patients (18). In contrast to TRITON II, TRITON III is a phase III randomized control trial evaluating the efficacy of rucaparib vs. abiraterone, enzalutamide, and docetaxel in patients with a deleterious mutation in BRCA1, BRCA2 or ATM. Other trials using PARP inhibitors are summarized in Table 2. Importantly, whether a single loss of function mutation is sufficient to confer sensitivity to PARP inhibitors or platinum therapy, or whether loss of the second allele (either through a bi-allelic mutation or loss of heterozygosity), is required for treatment response remains to be determined. As clinicians begin to utilize tumor sequencing more routinely to guide patient management, we will also undoubtedly uncover more variants of unknown significance (VUS) in these DDR genes. As not all mutations within a gene have the same functional consequence or impact on treatment sensitivity, we anticipate that careful annotation of these VUS and cross-referencing with established databases (e.g., ClinVar) will be required to best identify the patients most likely to benefit from these targeted approaches. The current clinical trials will undoubtedly help to answer some of these issues, and generate further hypotheses to test in preclinical models.
Full table
Response and resistance to PARP inhibitors
Unfortunately, most responses to PARP inhibitors are not sustained, and last only approximately 10–18 months (24,45), after which resistance develops. Mutations in PARP itself, the target of the PARP inhibitors, are not commonly found, although the protein levels of PARP and another protein, 53BP1, have been implicated in mediating resistance. In breast and ovarian cancer, BRCA2 reversion mutations that restore the translated reading frame have been shown to be a primary resistance mechanism to PARP inhibitors (46,47). In an analysis of patients from the TOPARP trial, de Bono and colleagues observed that reversion mutations could also be identified in cell-free DNA (cfDNA) in both patients with deleterious germline and somatic BRCA2 mutations (48). Our group has also identified BRCA2 reversion mutations in PARP inhibitor treated CRPC patients. Interestingly, these mutations were identified in cfDNA before clinical evidence of progression and resistance to PARP inhibitor therapy was observed, pointing towards the possibility of monitoring PARP sensitivity through the use of serial cfDNA sampling (49). At the University of California, San Francisco (UCSF), we are currently conducting a study sequencing the cfDNA of prostate cancer patients on any PARP inhibitor to characterize the spectrum of mutations that emerge prior to clinical detection of resistance. Similar to the development of PARP inhibitor resistance, BRCA2 reversion mutations may also represent a mechanism of resistance patients treated with platinum therapies, as was recently described in a case report published by Pritchard and colleagues (50). Taken together, we predict that BRCA reversions (and likely other biomarkers) will need to be carefully monitored by serially analyzing cfDNA in patients on single-agent PARP inhibitors or platinum therapies, which may herald the beginning the resistance and trigger the clinician to change therapy.
What about trying to prevent or delay resistance by using drug combinations that target multiple pathways? For example, targeting WEE1, a tyrosine kinase that inactivates CDK1/CDK2 in response to DNA damage, has not only been shown to have activity in BRCA-mutated cancer (51) but the combination with PARP inhibitors has also been shown in preclinical pancreatic cancer models to sensitize tumor cells to radiotherapy (52). Other strategies may involve utilizing alternating cycles of PARP inhibitor with taxane therapy, which may help to clear the PARP inhibitor-resistant clones. However, more preclinical studies and clinical trials will be required to determine whether rationale combination strategies will delay the emergence of PARP inhibitor resistance, and whether the side effect profile can be tolerated by patients.
AR signaling and DDR
AR plays a central role in prostate cancer, both in the hormone-sensitive and castrate-resistant settings (53). In preclinical models of prostate cancer, AR signaling is implicated in DNA damage and DDR (54-58). Androgen signaling promotes TOP2B mediated double-strand breaks, and the co-localization of AR and TOP2B to sites of TMPRSS2-ERG genomic breakpoints is essential for the generation of TMPRSS2-ERG rearrangements, which are present in more than 50% of prostate cancers (59). These rearrangements result from the fusion of the AR-responsive TMPRSS2 promoter with the ERG transcription factor, which is considered a quintessential driver of aggressive prostate cancer (60,61).
Interestingly, PARP1 appears to be an important contributor to the tumorigenic effects of TMPRSS2-ERG. PARP1 enhances the transcriptional function of AR, is required for tumor growth in xenograft models, and promotes the development of castration resistance (62). PARP1 interacts directly with ERG, and is required for the full activity of ERG. Additionally, ERG-positive xenografts are more sensitive to PARP inhibition (63). These observations led to a clinical trial by Hussain and colleagues to evaluate the efficacy of veliparib plus abiraterone vs. abiraterone monotherapy in patients with metastatic CRPC. The investigators hypothesized that ETS-fusion positive tumors would have an enhanced response to veliparib plus abiraterone compared to abiraterone alone. While there was no difference observed in the PSA response rate between the patients with ETS-fusion positive and negative tumors, the 25% of patients with HR deficiencies in both arms of the study had better responses than the patients with HR wildtype tumors. Progression free survival in patients with HR deficient tumors was 13.8 vs. 8.1 months in patients with HR wildtype tumors (P=0.0472). HR deficiency was also associated with a significantly better PSA rate response (90% vs. 56.7%; P=0.007) (64). In contrast, a study published by Annala et al., which included a cohort of 319 patients who had progressed to metastatic CRPC, demonstrated that DNA repair defects were associated with a shorter median time to castration resistance after initiating ADT (11.8 vs. 19.0 months, P=0.031) and median time to progression on abiraterone or enzalutamide (3.3 vs. 6.2 months, P=0.01) (65,66). The difference between the Annala et al. and the Hussain et al. analyses may be accounted for by the fact that Annala et al. looked at mutations present in the cfDNA of patients treated with either abiraterone or enzalutamide, while Hussain et al. looked at germline and somatic mutations present in primary tumor tissue in patients treated with abiraterone ± veliparib. Given the differences in trial design and the conflicting results, further investigation is needed to determine the prognostic implications of HR deficiency in the setting of treatment with second-generation anti-androgen therapies.
MMR deficiencies and immunotherapy in prostate cancer
In addition to homologous repair, cells have another DDR system, the MMR pathway, to correct base-base mismatches and insertion-deletion loops, which can also occur during the replication phase of the cell cycle (Figure 1). In tumors lacking MMR, long tracks of repeated sequences known as microsatellites are prone to strand slippage, which results in the insertion-deletion loops and the accumulation of mutations in these microsatellite areas. The classic example of MMR deficiency is Lynch syndrome, which is characterized by the germline loss of one of the canonical MMR genes (MLH1, MSH2, MSH6 and PMS2), and increases the risk of developing colorectal, endometrial and ovarian cancer, as well as prostate cancer (67). Cheng and colleagues previously identified deleterious germline MMR mutations in a small percentage of patients with metastatic prostate cancer (4 of the 692 patients, 0.6%) (18), suggesting that this is not a major pathway contributing to prostate cancer development. However, the combined prevalence of germline and somatic mutations are reportedly higher (approximately 3–12%, depending on the study) (68,69). Interestingly, hypermutated prostate cancers are characterized by complex MSH2 and MSH6 structural rearrangements, in contrast to colon cancer, which is often due to MLH1 epigenetic silencing or inactivating mutations (69). Next generation sequencing (NGS) assays that typically only sequence the exons of target genes may actually miss these MMR gene alterations resulting from intronic or non-coding region rearrangements because they do not typically capture these structural rearrangements. An alternative approach is to use immunohistochemistry (IHC) to detect MMR protein loss. Guedes and colleagues recently used IHC to screen >1,100 prostate cancer patients for MSH2 loss, and found that although MSH2 deficiency was rare in the general cohort (1%), it was enriched in patients with primary Gleason pattern 5 disease (8%) and small cell prostate cancer (5%) (70). This suggests that standard IHC may be an appropriate method to identify prostate cancer patients with MMR deficiency.
The loss of MMR is often associated with an increased mutational load, which is thought to result in increased tumor neoantigens due to changes in the amino acid sequence encoding proteins, which can be presented to immune system and recognized as non-self. This, in turn, has been hypothesized to augment the response to immune checkpoint inhibitors such as pembrolizumab, nivolumab and atezolimumab (71,72). A pivotal study by Le and colleagues showed that responses to anti-programmed death 1 (anti-PD-1) therapy were significantly better in patients with MMR deficiency compared to those without MMR deficiency (40% vs. 0%). This has led to the FDA approval of the anti-PD1 agent pembrolizumab in the treatment of patients with unresectable or metastatic MSI-high or MMR-deficiency solid tumors, the first FDA approved cancer therapy irrespective of the tissue of origin. Responders to checkpoint inhibitors may also include those without MMR deficiency or microsatellite instability. Indeed, in a small study of metastatic CRPC patients treated with pembrolizumab, one of the responder patients did not have detectable microsatellite instability (73). Additional work will be required to determine what other subtypes of prostate cancer, besides those with MMR deficiency or microsatellite instability, might benefit from this therapeutic approach. Of particular interest is whether DDR-deficient tumors are also immunoresponsive, which is currently being tested in clinical trials (Table 3). Moreover, for MMR-deficient tumors that do not respond to anti-PD1 agents, the combination with ipilimumab, an anti-cytotoxic T-lymphocyte associated protein 4 (CTLA4) monoclonal antibody, may also be considered.

Full table
Finally, there has also been considerable interest in combining anti-PD-1/PD-L1 agents with other novel agents to stimulate immunotherapy responses in patients without MMR deficiency. Given that PARP inhibitors may be able to trigger genomic instability, and that tumors with HR deficiencies are already unstable, there has been interest in combining immunotherapy with PARP inhibition in advanced prostate cancer. An ongoing study by Karzai and colleagues using the combination of durvalumab (an anti-PD-L1 agent) and olaparib reported that approximately 40% of patients unselected for HR or MMR deficiency had a documented decline in their PSA. In addition, studies combining nivolumab plus another PARP inhibitor, rucaparib, are also underway, as are immunotherapy combinations with conventional taxane chemotherapy in urothelial carcinoma (i.e., KEYNOTE-395). In addition, novel agents in combination with checkpoint inhibition are being explored as well. For example, inhibitors of indoleamine dioxygenase (IDO), an enzyme involved in the metabolomic checkpoint, in combination with anti-PD1 therapy are in clinical trials for multiple tumor types, although recent data in melanoma from the ECHO-301/KEYNOTE-252 trial were disappointing. Whether these combinations in particular molecular subtypes of prostate cancer will enhance immunotherapy sensitivity remains to evaluated, and is an exciting area of ongoing investigation.
Future outlook and conclusions
Over the last several years, we have gained a deeper understanding of the molecular alterations that define prostate cancer. Surprisingly, one of the key hallmarks that emerged from the multiple large-scale sequencing studies was that a significant subset of prostate cancer involves mutations in genes involved in various DNA repair pathways, including HR and MMR. These insights have led to the study and use of new agents to treat prostate cancer, including PARP inhibitors, platinum chemotherapy, and checkpoint immunotherapy. It is important to remember that several other DNA repair pathways exist (e.g., NHEJ), which are regulated and executed by a suite of other unique enzymes, all of which may be potential targets for therapy. This includes drugs targeting DNAPK and ATR, which are currently under clinical testing and development.
With these paradigm shifts occurring in prostate cancer treatment, understanding the genomic features of each individual patient’s tumor is becoming increasingly important. Choosing the right treatment for the right patient and integrating genomic information with clinical data to inform treatment decisions continue to be the goal of precision oncology. One outstanding question is whether homozygous mutations or deletions are required for sensitivity to these targeted approaches, or whether hemizygous mutations are sufficient for therapeutic response. In considering HR deficiency, genes such as BRCA1, BRCA2, and ATM are classically considered tumor suppressors. As such it would be expected that loss of both copies would be required for therapeutic sensitivity. However, there is currently a paucity of conclusive data supporting this hypothesis. But as suggested in the TOPARP-A trial, the vast majority of PARP inhibitor responders had homozygous loss of function HR gene mutations or deletions. Ongoing trials such as TRITON II/III only require a single-copy mutation for eligibility, and so this will be a critical issue to address. Similarly, as the field of precision oncology continues to mature, the annotation of VUS will be a crucial element in the development of targeted therapies. To this end, it will be important to have robust functional assays and preclinical models to help predict and evaluate which variants are likely to confer response.
At the same time, we must continue to understand treatment resistance to stay a few steps ahead of the tumor and be able to provide effective treatment options for the patient. This will involve not only setting up the pipeline to perform sequential biopsies of metastatic lesions, but also the pipeline to interrogate the whole genome, exome, transcriptome, and proteome in order to understand the multiple pathways to resistance. In addition, we need to improve the technology to sequence cfDNA and capture circulating tumor cells, or CTCs, in order to allow us to obtain serial liquid biopsies, which are particularly useful in cases where biopsies may not be possible (e.g., of metastatic spine lesions or abdominal lymph nodes). The incorporation of omics in prostate cancer will open new therapeutic opportunities for patients and new areas of investigation for clinical, translational and basic scientists, which may ultimately allow oncologists to achieve cures for patients with metastatic, castrate-resistant disease.
Acknowledgements
J Chou is supported by a T32 training grant from the NCI (CA108462-13S1) and funds from the Quinlan Fellowship in Genitourinary Oncology.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137-50. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Beer TM, Armstrong AJ, Rathkopf DE, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371:424-33. [Crossref] [PubMed]
- Ryan CJ, Cheng ML. Abiraterone acetate for the treatment of prostate cancer. Expert Opin Pharmacother 2013;14:91-6. [Crossref] [PubMed]
- Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 2010;363:411-22. [Crossref] [PubMed]
- Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004;351:1502-12. [Crossref] [PubMed]
- de Bono JS, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet 2010;376:1147-54. [Crossref] [PubMed]
- Parker C, Nilsson S, Heinrich D, et al. Alpha Emitter Radium-223 and Survival in Metastatic Prostate Cancer. N Engl J Med 2013;369:213-23. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215-28. [Crossref] [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [Crossref] [PubMed]
- Beltran H, Yelensky R, Frampton GM, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol 2013;63:920-6. [Crossref] [PubMed]
- Beltran H, Rickman DS, Park K, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 2011;1:487-95. [Crossref] [PubMed]
- Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298-305. [Crossref] [PubMed]
- Bluemn EG, Coleman IM, Lucas JM, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017;32:474-89.e6. [Crossref] [PubMed]
- Zhao SG, Chang SL, Erho N, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol 2017;3:1663-72. [Crossref] [PubMed]
- Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol 2016;17:1612-20. [Crossref] [PubMed]
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375:443-53. [Crossref] [PubMed]
- Prakash R, Zhang Y, Feng W, et al. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 2015;7:a016600. [Crossref] [PubMed]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is Required for Homology-Directed Repair of Chromosomal Breaks. Molecular Cell 2001;7:263-72. [Crossref] [PubMed]
- Xia F, Taghian DG, DeFrank JS, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. PNAS 2001;98:8644-9. [Crossref] [PubMed]
- Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 2001;20:4704-16. [Crossref] [PubMed]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012;22:106-16. [Crossref] [PubMed]
- Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 2015;373:1697-708. [Crossref] [PubMed]
- Leongamornlert D, Mahmud N, Tymrakiewicz M, et al. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer 2012;106:1697-701. [Crossref] [PubMed]
- Kote-Jarai Z, Leongamornlert D, Saunders E, et al. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer 2011;105:1230-4. [Crossref] [PubMed]
- Tryggvadottir L, Vidarsdottir L, Thorgeirsson T, et al. Prostate cancer progression and survival in BRCA2 mutation carriers. J Natl Cancer Inst 2007;99:929-35. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. [Crossref] [PubMed]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110-20. [Crossref] [PubMed]
- D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 2003;3:23-34. [Crossref] [PubMed]
- Venkitaraman AR. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer 2004;4:266-76. [Crossref] [PubMed]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature 2012;481:287-94. [Crossref] [PubMed]
- Turner NC, Tutt AN. Platinum chemotherapy for BRCA1-related breast cancer: do we need more evidence? Breast Cancer Res 2012;14:115. [Crossref] [PubMed]
- Lohse I, Borgida A, Cao P, et al. BRCA1 and BRCA2 mutations sensitize to chemotherapy in patient-derived pancreatic cancer xenografts. Br J Cancer 2015;113:425-32. [Crossref] [PubMed]
- Byrski T, Dent R, Blecharz P, et al. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res 2012;14:R110. [Crossref] [PubMed]
- Pomerantz MM, Spisák S, Jia L, et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 2017;123:3532-9. [Crossref] [PubMed]
- Sternberg CN, Whelan P, Hetherington J, et al. Phase III trial of satraplatin, an oral platinum plus prednisone vs. prednisone alone in patients with hormone-refractory prostate cancer. Oncology 2005;68:2-9. [Crossref] [PubMed]
- Buonerba C, Federico P, Bosso D, et al. Carboplatin plus etoposide in heavily pretreated castration-resistant prostate cancer patients. Future Oncol 2014;10:1353-60. [Crossref] [PubMed]
- Vaishampayan U, Shevrin D, Stein M, et al. Phase II Trial of Carboplatin, Everolimus, and Prednisone in Metastatic Castration-resistant Prostate Cancer Pretreated With Docetaxel Chemotherapy: A Prostate Cancer Clinical Trial Consortium Study. Urology 2015;86:1206-11. [Crossref] [PubMed]
- Kentepozidis N, Soultati A, Giassas S, et al. Paclitaxel in combination with carboplatin as salvage treatment in patients with castration-resistant prostate cancer: a Hellenic oncology research group multicenter phase II study. Cancer Chemother Pharmacol 2012;70:161-8. [Crossref] [PubMed]
- Ross RW, Beer TM, Jacobus S, et al. A phase 2 study of carboplatin plus docetaxel in men with metastatic hormone-refractory prostate cancer who are refractory to docetaxel. Cancer 2008;112:521-6. [Crossref] [PubMed]
- Hager S, Ackermann CJ, Joerger M, et al. Anti-tumour activity of platinum compounds in advanced prostate cancer—a systematic literature review. Ann Oncol 2016;27:975-84. [Crossref] [PubMed]
- Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949-61. [Crossref] [PubMed]
- Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008;451:1111-5. [Crossref] [PubMed]
- Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008;451:1116-20. [Crossref] [PubMed]
- Goodall J, Mateo J, Yuan W, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov 2017;7:1006-17. [Crossref] [PubMed]
- Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of Circulating Cell-Free DNA Identifies Multiclonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov 2017;7:999-1005. [Crossref] [PubMed]
- Cheng HH, Salipante SJ, Nelson PS, et al. Polyclonal BRCA2 Reversion Mutations Detected in Circulating Tumor DNA After Platinum Chemotherapy in a Patient With Metastatic Prostate Cancer. JCO Precision Oncology 2018. doi: 10.1200/PO.17.00169. [Crossref]
- Do K, Wilsker D, Ji J, et al. Phase I Study of Single-Agent AZD1775 (MK-1775), a Wee1 Kinase Inhibitor, in Patients With Refractory Solid Tumors. J Clin Oncol 2015;33:3409-15. [Crossref] [PubMed]
- Karnak D, Engelke CG, Parsels LA, et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res 2014;20:5085-96. [Crossref] [PubMed]
- Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013;32:5501-11. [Crossref] [PubMed]
- Ta HQ, Gioeli D. The convergence of DNA damage checkpoint pathways and androgen receptor signaling in prostate cancer. Endocr Relat Cancer 2014;21:R395-407. [Crossref] [PubMed]
- Tarish FL, Schultz N, Tanoglidi A, et al. Castration radiosensitizes prostate cancer tissue by impairing DNA double-strand break repair. Sci Transl Med 2015;7:312re11. [Crossref] [PubMed]
- Polkinghorn WR, Parker JS, Lee MX, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov 2013;3:1245-53. [Crossref] [PubMed]
- Schiewer MJ, Knudsen KE. Linking DNA Damage and Hormone Signaling Pathways in Cancer. Trends Endocrinol Metab 2016;27:216-25. [Crossref] [PubMed]
- Goodwin JF, Schiewer MJ, Dean JL, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 2013;3:1254-71. [Crossref] [PubMed]
- Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 2010;42:668-75. [Crossref] [PubMed]
- Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 2006;66:8337-41. [Crossref] [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [Crossref] [PubMed]
- Schiewer MJ, Goodwin JF, Han S, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov 2012;2:1134-49. [Crossref] [PubMed]
- Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell 2011;19:664-78. [Crossref] [PubMed]
- Hussain M, Daignault-Newton S, Twardowski PW, et al. Targeting Androgen Receptor and DNA Repair in Metastatic Castration-Resistant Prostate Cancer: Results From NCI 9012. J Clin Oncol 2018;36:991-9. [Crossref] [PubMed]
- Annala M, Struss WJ, Warner EW, et al. Treatment Outcomes and Tumor Loss of Heterozygosity in Germline DNA Repair–deficient Prostate Cancer. Eur Urol 2017;72:34-42. [Crossref] [PubMed]
- Chi KN, Annala M, Sunderland K, et al. A randomized phase II cross-over study of abiraterone + prednisone (ABI) vs enzalutamide (ENZ) for patients (pts) with metastatic, castration-resistant prostate cancer (mCRPC). J Clin Oncol 2017;35:5002. [Crossref]
- Raymond VM, Mukherjee B, Wang F, et al. Elevated risk of prostate cancer among men with Lynch syndrome. J Clin Oncol 2013;31:1713-8. [Crossref] [PubMed]
- Schweizer MT, Cheng HH, Tretiakova MS, et al. Mismatch repair deficiency may be common in ductal adenocarcinoma of the prostate. Oncotarget 2016;7:82504-10. [Crossref] [PubMed]
- Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun 2014;5:4988. [Crossref] [PubMed]
- Guedes LB, Antonarakis ES, Schweizer MT, et al. MSH2 Loss in Primary Prostate Cancer. Clin Cancer Res 2017;23:6863-74. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Kelderman S, Schumacher TN, Kvistborg P. Mismatch Repair-Deficient Cancers Are Targets for Anti-PD-1 Therapy. Cancer Cell 2015;28:11-3. [Crossref] [PubMed]
- Graff JN, Alumkal JJ, Drake CG, et al. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 2016;7:52810-7. [Crossref] [PubMed]