
Deep neural networks could differentiate Bethesda class III versus class IV/V/VI
Introduction
Thyroid nodules are common. Studies (1) have shown an incidence of 19–35% on ultrasound (US) and up to 65% incidence at autopsy. US can provide high-resolution images of thyroid gland.
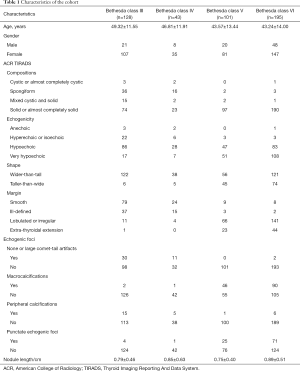
ACR (American College of Radiology) Thyroid Imaging Reporting And Data System (TIRADS) is a widely used risk stratification system evaluating the benignity/malignancy of thyroid nodules (2). ACR TIRADS quantifies five categories of US features and assigns different scores to each category. The five categories are composition (cystic or almost completely cystic, spongiform, mixed cystic and solid, solid or almost completely solid), echogenicity (anechoic, hyperechoic or isoechoic, hypoechoic, very hypoechoic), shape (wider-than-tall, taller-than-wide), margin (smooth, ill-defined, lobulated or irregular, extra-thyroidal extension), and echogenic foci (none or large comet-tail artifacts, macrocalcifications, peripheral calcifications, punctate echogenic foci).
The fine needle aspirate is most commonly classified using the Bethesda System for Reporting Thyroid Cytology (TBSRTC) (3). Studies have shown that TBSRTC provide adequate communication between pathologists and radiologists with high accuracy (98%) and low false positive rate (3%) (4). However, studies show that TIRADS classification has only 70% concordance with the Bethesda grade (5). This is clinically relevant because different Bethesda grades have different treatments. Malignant nodules (Bethesda class V/V/VI) are surgically resected while benign nodules (Bethesda class II) are followed. Bethesda class III nodules may require an additional biopsy to provide material for genetic analysis to determine appropriate treatment. More accurate characterization of the nodules by US prior to the biopsy may thus be helpful in clinical management.
In this paper, we defined our problem as a binary classification problem. Our goal was to differentiate Bethesda class III from Bethesda grade IV/V/VI nodules using machine learning and deep learning approaches. We applied logistic regression, random forest, support vector machine (SVM), gradient boosting, k-nearest neighbors (KNN) and deep neural networks (DNN) for this binary classification.
Methods
A retrospective study of 467 thyroid FNAs conducted by the Department of Pathology at the Sichuan Cancer Hospital & Institute from March 2017 to March 2018 was undertaken. US images and corresponding features, demographic information, and cytopathology results were collected and described. The nodules were described by an experienced radiologist using the TIRADS lexicon. US images were collected using Phillips, GE, Siemens and Supersonic US machines.
All FNAs were performed using US guidance to directly visualize the needle tip within the nodule of interest. FNAB was performed primarily with 27-gauge needles and capillary action without a syringe or aspirator. The needle was attached to an air-filled syringe to express the specimen on a glass slide. A second glass slide was used to smear the specimen into a thin layer, creating two direct smears from each pass. One smear was immediately submerged in alcohol for Papanicolaou stain. The FNAs were evaluated by two pathologists with more than 5 years of cytopathology experience.
Six supervised classification methods (logistic regression, random forest, decision tree, gradient boosting, SVM, KNN, DNN) were tested on the cohort. The Bethesda class III cases were labeled as class “0”, while the malignant Bethesda class VI/V/VI, were labeled as class “1”. We randomly chose 75% of the data as the training set, 10% as the validation set, and 15% as the test set. The six classifiers were built for this binary classification task. Cross validations with number of five folds were conducted when testing the model.
Results
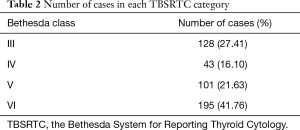
A total of 467 cytopathological cases were collected in the Department of Pathology. The cohort was comprised of 370 female patients (female to male ratio 3.8:1), age varied from 9 to 83 years (45.31±13.28 years) and nodule length varied from 0.2 to 3.5 cm (0.83±0.49 cm). Table 1 shows the number of cases for each ACR TIRADS feature. Table 2 demonstrates the number of cases in each TBSRTC category.
Full table
Full table
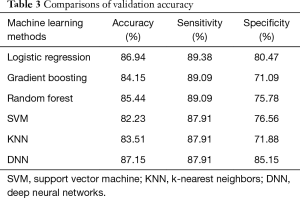
DNN achieved the highest accuracy of 87.15%. Other classifiers achieved from 82.23% (SVM) to 86.94% (logistic regression) accuracy. The detailed comparisons of the models’ performance are shown in Table 3.
Full table
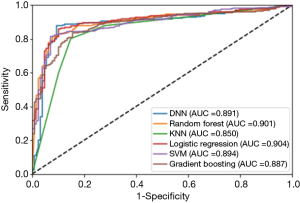
Figure 1 indicates the receiver operating curve (ROC) of the six classifiers. Area under curve (AUC) is an indicator to show the performance of each method. All approaches showed similar results in AUC. Logistic regression slightly outperformed other classifiers (AUC =0.904), compared to gradient boosting (AUC =0.887), random forest (AUC =0.901), SVM (AUC =0.894), KNN (AUC =0.850) and DNN (AUC =0.891).
Discussion
Machine learning and deep learning methods have previously been used to classify thyroid nodules using US images and/or radiologists’ description of the nodules. Wu et al. (6) built machine learning algorithms to differentiate suspicious thyroid nodules via sonography. Mei et al. (7) applied convolutional autoencoders associated with TIRADS descriptions to predict the benignity of thyroid nodules. Chang et al. (8) applied SVM to classify benign and malignant thyroid nodules based on US images. Gopinath et al. (9) integrated statistical texture features and FNA cytology microscopic images to an SVM classifier to classify benign and malignant thyroid nodules. These studies show machine learning approaches can achieve similar or higher accuracy than radiologists in differentiating benign and malignant nodules based on US images.
Most of the prior studies have focused on classifying benign and malignant nodules and achieved different accuracies based on different sizes of the datasets. Our study, however, applied machine learning tools to further classify suspicious nodules that might require different treatments.
Conclusions
As can be seen in Table 3 and Figure 1, our machine learning approaches are helpful in differentiating Bethesda class III from classes IV/V/VI. DNN, random forest, logistic regression and SVM are useful models both with respect to accuracy and AUC values. This could help radiologists and pathologists to better manage thyroid nodules and provide more effective and efficient treatment for Bethesda class III nodules.
Limitations of this study include nodule feature description by only one radiologist, possibly introducing bias. Further studies should include descriptions from multiple radiologists. In addition, the study used a relatively small number of nodules from a single hospital. Future studies could include additional data from additional sites.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Internal Ethical Committee at Sichuan Cancer Hospital and written informed consent was obtained from all subjects.
References
- Keh SM, El-Shunnar SK, Palmer T, et al. Incidence of malignancy in solitary thyroid nodules. J Laryngol Otol 2015;129:677-81. [Crossref] [PubMed]
- Tessler FN, Middleton WD, Grant EG, et al. ACR Thyroid Imaging, Reporting and Data System (TI-RADS): White Paper of the ACR TI-RADS Committee. J Am Coll Radiol 2017;14:587-95. [Crossref] [PubMed]
- Crowe A, Linder A, Hameed O, et al. The impact of implementation of the Bethesda System for Reporting Thyroid Cytopathology on the quality of reporting, “risk” of malignancy, surgical rate, and rate of frozen sections requested for thyroid lesions. Cancer Cytopathol 2011;119:315-21. [Crossref] [PubMed]
- Renuka IV, Saila Bala G, Aparna C, et al. The bethesda system for reporting thyroid cytopathology: interpretation and guidelines in surgical treatment. Indian J Otolaryngol Head Neck Surg 2012;64:305-11. [Crossref] [PubMed]
- Vargas-Uricoechea H, Meza-Cabrera I, Herrera-Chaparro J. Concordance between the TIRADS ultrasound criteria and the BETHESDA cytology criteria on the nontoxic thyroid nodule. Thyroid Res 2017;10:1. [Crossref] [PubMed]
- Wu H, Deng Z, Zhang B, et al. Classifier Model Based on Machine Learning Algorithms: Application to Differential Diagnosis of Suspicious Thyroid Nodules via Sonography. AJR Am J Roentgenol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Mei X, Dong X, Deyer T, et al. Thyroid Nodule Benignity Prediction by Deep Feature Extraction. Washington, DC: 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE), 2017:241-5.
- Chang Y, Paul AK, Kim N, et al. Computer-aided diagnosis for classifying benign versus malignant thyroid nodules based on ultrasound images: A comparison with radiologist-based assessments. Med Phys 2016;43:554. [Crossref] [PubMed]
- Gopinath B, Shanthi N. Support Vector Machine based diagnostic system for thyroid cancer using statistical texture features. Asian Pac J Cancer Prev 2013;14:97-102. [Crossref] [PubMed]


