
ABCA7 genotype altered Aβ levels in cerebrospinal fluid in Alzheimer’s disease without dementia
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by early short-term memory loss progressing into dementia. It has been documented that clinical symptoms result from the deposition of β-amyloid (Aβ) and tau pathology in brain, and Aβ has been observed to mainly aggregate and accumulate in the brain in non-dementia AD until a critical threshold is reached in AD, which exhibits a sigmoidal trajectory characterized by a rapid progression and finally a plateau. Aβ deposition can be measured by cortical amyloid positron emission tomography (PET) ligand binding (1,2) or low cerebrospinal fluid (CSF) Aβ1–42 (3-5).
ATP-binding cassette, subfamily A, member 7 (ABCA7) has been found to be expressed abundantly in brain microglia, implying a role of ABCA7 in amyloid clearance and fibril formation (1-5). ABCA7 loss-of-function (LOF) was reported to contribute to AD by promoting Aβ processing and pathology in cell culture and mouse models (1). In addition, our group identified that ABCA7 variants were associated with Aβ deposition on amyloid imaging but were not related to Aβ levels in CSF test (6). Recently, ABCA7 variable number tandem repeat (VNTR) length was observed to correlate with CSF Aβ1–42 (5). Therefore, the relation between ABCA7 polymorphisms and CSF Aβ1–42 was still inconsistent across these studies, and it’s necessary to further identify the association, especially in the non-dementia AD stage in which Aβ is mostly deposited.
To date, ABCA7 rs3764650 has been identified to be significantly associated with AD susceptibility in genome-wide association studies (GWAS) and following studies (7-18). To explore the functional consequences of the rs3764650 in ABCA7 and its possible effect on AD pathogenesis, we attempted to analyze its associations with CSF proteins (Aβ1–42, t-tau, and p-tau) in non-dementia AD subjects.
Methods
ADNI dataset
ADNI was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies and nonprofit organizations (http://adni.loni.usc.edu). ADNI, with Michael W. Weiner, MD as the principal investigator, was approved by the regional ethical committees of all participating institutions and written informed consent was obtained from all participants.
Subject classification
Subjects were classified into preclinical and prodromal AD by CSF Aβ1–42 or 18FAV-45-PET according to the National Institute on Aging and Alzheimer’s Association (NIA-AA) research criteria in 2011. The cutoff value for abnormal CSF Aβ1–42 levels was ≤192 pg/mL or 18FAV-45-PET ≥1.11. The application of a normal/abnormal cut point for each amyloidosis biomarker divided the non-dementia subjects into amyloid group (non-dementia Aβ+) and non-amyloid group (non-dementia Aβ−) and subdivided the former into preclinical AD and prodromal AD from CN and mild cognitive impairment (MCI) group respectively.
Genotyping
ADNI samples were genotyped using the Illumina Human 610-Quad Bead Chip array (Illumina, Inc., San Diego, CA, USA), including 620,901 SNP and CNV markers which were employed for genotyping. Here we extracted the genetic data of ABCA7 (rs3764650) from the ADNI PLINK data format.
CSF proteins
Methods for the collection, extraction, and examination of CSF proteins, including Aβ1–42, total tau (t-tau), and phosphorylated tau (p-tau), have been described previously (19,20). Finally, a total of 410 non-dementia Aβ+ subjects (104 preclinical AD subjects and 306 prodromal AD subjects) were included in CSF analysis from the ADNI sites.
Data analysis
All statistical analyses were conducted using R. We used a multiple linear regression model to analyze the effects of ABCA7 rs3764650 on CSF biomarkers at baseline with corrections for age, gender, educational level, and APOE ε4 genotype; and we used a linear mixed effects model (including age, gender, educational level, and APOE ε4 genotype as covariates) with random intercepts and slope for time to test the association of rs3764650 with the levels of CSF biomarkers from longitudinal data (21). For all the tests, we claim that ABCA7 rs3764650 is significantly associated with the levels of CSF biomarkers when P values are below 0.05. Moreover, our subjects were restricted to non-Hispanic and white people to avoid genetic stratification across ethnicities.
Results
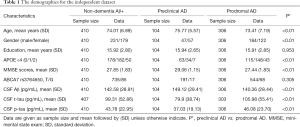
Finally, 410 non-dementia Aβ+ subjects (231 male vs. 179 female) consisted of 104 preclinical AD subjects (47 male vs. 57 female) and 306 prodromal AD subjects (184 male vs. 122 female). The demographic characteristics and distributions of the genotypes are shown in Table 1. Prodromal AD had the higher frequency for the APOE ε4 allele and performed worse than did preclinical AD on MMSE. As expected, the levels of CSF Aβ1–42, t-tau and p-tau were different between the two groups, and prodromal AD had lower CSF Aβ1–42, higher t-tau and p-tau levels than did preclinical AD. We used the dominant model in our analysis because the number of homozygotes (GG) was too small.
Full table
Finally, GG + GT genotypes (77, 138.38±31.95 pg/mL) showed lower Aβ1–42 level when compared to TT genotype (333, 143.55±28.12 pg/mL) in the non-dementia Aβ+ group. Moreover, GG + GT genotypes (61, 135.11±30.81 pg/mL) had lower Aβ1–42 level than did TT genotypes (245, 141.66±27.73 pg/mL) in the prodromal AD group, while they did not differ in CSF Aβ1–42 level in the preclinical AD group. In the multiple linear regression model, GG + GT genotypes significantly decreased the level of CSF Aβ1–42 in non-dementia Aβ+ group (β=−8.8043, P<0.05), and a negative correlation between rs3764650 polymorphisms and CSF Aβ1–42 was also found in the prodromal AD group (β=−9.9756, P<0.01) (Figure 1). No significant relationships between the levels of CSF t-tau and p-tau and ABCA7 rs3764650 were observed at baseline in all diagnostic groups (P>0.05) (Table 2).


Full table
Furthermore, we used a linear mixed effects model to test this association in longitudinal studies. The significant negative association of the rs3764650 polymorphism with CSF Aβ1–42 was also present within non-dementia Aβ+ group (β=−0.3328, P<0.005) and within prodromal AD group (β=−0.3567, P<0.01) (Figure 2). However, the associations of rs3764650 with other CSF proteins (t-tau and p-tau) were also not detected in longitudinal studies (P>0.05) (Table 3).


Full table
Discussion
This study provided a comprehensive evaluation of the impact of ABCA7 SNP rs3764650 on CSF proteins (Aβ1–42, t-tau and p-tau) in non-dementia AD, including preclinical AD and prodromal AD. In non-dementia AD, rs3764650 was detected to correlate with low CSF Aβ1–42 level rather than CSF tau proteins (t-tau and p-tau) at baseline, and the correlation between ABCA7 and CSF Aβ1–42 was further verified in the prodromal AD group. As we further extended these findings in longitudinal studies, the significant associations still existed in non-dementia AD and prodromal AD. Therefore, our study detected that ABCA7 increased AD risk through involving in Aβ deposition but not in tau pathology, which was consistent with the previous findings.
ABCA7 was identified as a known risk factor for AD in genetic analyses. Former studies have found that ABCA7 variants contributed to AD by altering the clearance of microglia or the phagocytosis of macrophages, resulting in Aβ accumulation (22-27). In addition, ABCA7 variants have been observed to be associated with Aβ deposition on amyloid imaging, but not associated with the level of Aβ1–42 in CSF (6). Different from previous studies, our study explored the correlation between ABCA7 and the CSF Aβ1–42 level in the non-dementia AD stage (from preclinical AD to prodromal AD), which may explain the inconsistency between our result and those of other studies.
Conclusions
In summary, our study showed that ABCA7 genetic variants altered CSF Aβ1–42 level in non-dementia AD to modify the AD risk, which provided new evidence for the hypothesis that ABCA7 may contribute to the AD risk by altering Aβ deposition rather than tau pathology.
Acknowledgements
Funding: Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This work was also supported by grants from the National Key R&D Program of China (2016YFC1305803), the National Natural Science Foundation of China (81701253), Qingdao Key Health Discipline Development Fund, Qingdao Outstanding Health Professional Development Fund, and Shandong Provincial Collaborative Innovation Center for Neurodegenerative Disorders.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: ADNI, with Michael W. Weiner, MD as the principal investigator, was approved by the regional ethical committees of all participating institutions and written informed consent was obtained from all participants.
References
- Satoh K, Abe-Dohmae S, Yokoyama S, et al. ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. J Biol Chem 2015;290:24152-65. [Crossref] [PubMed]
- Vasquez JB, Fardo DW, Estus S. ABCA7 expression is associated with Alzheimer's disease polymorphism and disease status. Neurosci Lett 2013;556:58-62. [Crossref] [PubMed]
- Kim WS, Li H, Ruberu K, et al. Deletion of Abca7 increases cerebral amyloid-beta accumulation in the J20 mouse model of Alzheimer's disease. J Neurosci 2013;33:4387-94. [Crossref] [PubMed]
- Chan SL, Kim WS, Kwok JB, et al. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem 2008;106:793-804. [Crossref] [PubMed]
- De Roeck A, Duchateau L, Van Dongen J, et al. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease. Acta Neuropathol 2018;135:827-37. [Crossref] [PubMed]
- Zhao QF, Wan Y, Wang HF, et al. ABCA7 Genotypes Confer Alzheimer's Disease Risk by Modulating Amyloid-beta Pathology. J Alzheimers Dis 2016;52:693-703. [Crossref] [PubMed]
- Sims R, van der Lee SJ, Naj AC, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet 2017;49:1373-84. [Crossref] [PubMed]
- Moreno DJ, Ruiz S, Rios A, et al. Association of GWAS Top Genes With Late-Onset Alzheimer's Disease in Colombian Population. Am J Alzheimers Dis Other Demen 2017;32:27-35. [Crossref] [PubMed]
- Omoumi A, Fok A, Greenwood T, et al. Evaluation of late-onset Alzheimer disease genetic susceptibility risks in a Canadian population. Neurobiol Aging 2014;35:936.e5-12. [Crossref] [PubMed]
- Carrasquillo MM, Khan Q, Murray ME, et al. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurology 2014;82:1455-62. [Crossref] [PubMed]
- Tan L, Yu JT, Zhang W, et al. Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement 2013;9:546-53. [Crossref] [PubMed]
- Miyashita A, Koike A, Jun G, et al. SORL1 is genetically associated with late-onset Alzheimer's disease in Japanese, Koreans and Caucasians. PLoS One 2013;8:e58618. [Crossref] [PubMed]
- Chung SJ, Lee JH, Kim SY, et al. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis Assoc Disord 2013;27:250-7. [Crossref] [PubMed]
- Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436-41. [Crossref] [PubMed]
- Logue MW, Schu M, Vardarajan BN, et al. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol 2011;68:1569-79. [Crossref] [PubMed]
- Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088-93. [Crossref] [PubMed]
- Liu LH, Xu J, Deng YL, et al. A complex association of ABCA7 genotypes with sporadic Alzheimer disease in Chinese Han population. Alzheimer Dis Assoc Disord 2014;28:141-4. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol 2009;65:403-13. [Crossref] [PubMed]
- Olsson A, Vanderstichele H, Andreasen N, et al. Simultaneous measurement of beta-amyloid(1–42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin Chem 2005;51:336-45. [Crossref] [PubMed]
- Mattsson N, Andreasson U, Zetterberg H, et al. Association of Plasma Neurofilament Light With Neurodegeneration in Patients With Alzheimer Disease. JAMA Neurol 2017;74:557-66. [Crossref] [PubMed]
- Fleisher AS, Chen K, Quiroz YT, et al. Associations between biomarkers and age in the presenilin 1 E280A autosomal dominant Alzheimer disease kindred: a cross-sectional study. JAMA Neurol 2015;72:316-24. [Crossref] [PubMed]
- Fagan AM, Xiong C, Jasielec MS, et al. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer's disease. Sci Transl Med 2014;6:226ra30. [Crossref] [PubMed]
- Benzinger TL, Blazey T, Jack CR Jr, et al. Regional variability of imaging biomarkers in autosomal dominant Alzheimer's disease. Proc Natl Acad Sci U S A 2013;110:E4502-9. [Crossref] [PubMed]
- Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med 2012;367:795-804. [Crossref] [PubMed]
- Bennett DA, Schneider JA, Wilson RS, et al. Neurofibrillary tangles mediate the association of amyloid load with clinical Alzheimer disease and level of cognitive function. Arch Neurol 2004;61:378-84. [Crossref] [PubMed]
- Insel PS, Hansson O, Mackin RS, et al. Amyloid pathology in the progression to mild cognitive impairment. Neurobiol Aging 2018;64:76-84. [Crossref] [PubMed]


