A pathogenic hierarchy for synovial fibroblasts in rheumatoid arthritis
Rheumatoid arthritis (RA) is a chronic autoimmune disorder that affects 0.5–1% of the world’s population (1). It is characterized by inflammation and progressive destruction of joints (1). Synovial fibroblasts play important roles in initiating and driving RA by secreting inflammatory cytokines and chemokines, degrading cartilage, and stimulating osteoclasts that lead to bone erosion (2).
Importantly, synovial fibroblasts display altered phenotypes (3) and functions (4) in RA, due at least in part to changes in DNA methylation (5), making it plausible to target them therapeutically. Despite their fundamental importance, this goal has not yet been realised. Their molecular identity is still ambiguous, and diversities of phenotype and function are still topics of highly active study.
New work published in Nature Communications by Mizoguchi et al., takes an important step towards the development of an anti-synovial therapy for RA, and paves the way towards more meaningful methods of patient stratification and therapy evaluation (6). It reveals the first in-depth unbiased analyses of synovial fibroblast subsets freshly isolated from human tissues, and the subsequent assignment of a bioinformatics-driven pathogenic hierarchy of fibroblast subsets within the rheumatoid synovial microenvironment (6).
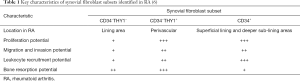
To investigate the heterogeneity of fibroblasts in joint tissue, Mizoguchi et al. compared uncultured, freshly isolated synovial fibroblasts from patients with RA or osteoarthritis (OA), to reveal autoimmune-specific changes. Flow cytometry identified seven distinct subsets of synovial fibroblasts based on expression of known surface markers including podoplanin (PDPN), THY1 (CD90), cadherin-11 (CDH11) and CD34. Utilising fluorescence activated cell sorting followed by transcriptomic profiling (microarray and RNA-Seq), these populations were analysed, both at the level of bulk-sorted subset and single cell, with functional follow-up (6).
Principal component analysis of sorted subsets, which provides an unbiased measure of heterogeneity, revealed that the seven distinct populations fell into three major subsets: CD34−THY1−, CD34–THY1+, and CD34+. This was independently supported by hierarchical clustering of RNA-Seq from single cells, where clustering based on 23 genes with greatest 1% mean and greatest 1% variance in expression levels was consistent with the major three subpopulations achieved by bulk transcriptomics of subsets gated by protein surface markers CD34 and THY1 (6).
The fibroblast subsets localised to distinct anatomical regions of the synovium, which may contribute to transcriptomic differences between RA and OA synovial tissue. In RA, the authors found that CD34−THY1+ synovial fibroblasts are enriched around blood vessels in the deep sub-lining area of the synovium, whereas in OA, the CD34−THY1+ subset formed a thin layer in the sub-lining area with fewer cells surrounding blood vessels. The increased proportion of CD34−THY1+ fibroblasts near the capillaries in the sub-lining layer differentiates RA from OA and reflects disease activity in RA (6).
Using flow cytometry and clinical evaluation of synovitis, the proportion of CD34−THY1+ fibroblasts among total fibroblasts significantly correlated with synovial hypertrophy and inflammation. Among RA patients, a comparison of swollen to non-swollen joints revealed a significantly higher proportion of CD34+ fibroblasts and CD34−THY1+ fibroblasts, and lower CD34−THY1– fibroblasts, suggesting that the CD34+ fibroblasts and CD34−THY1+ fibroblasts may play a stronger role in synovitis.
The authors therefore examined comparative expression of genes related to synoviocyte proliferation, invasion, and inflammation and followed this up by examining protein expression and functional assays.
Both CD34−THY1+ and CD34+ populations showed phenotypes suggestive of synoviocyte hypertrophy and invasion. Genes relevant to proliferation and cell cycle were comparatively upregulated in the CD34−THY1+ subset compared to CD34+ or double negative subsets, but in vitro experiments revealed that both CD34−THY1+ and CD34+ populations exhibited significantly higher proportions of Ki-67-positive (i.e., actively cycling) cells compared to the CD34−THY1− subset. Furthermore, the CD34−THY1+ and CD34+ subsets also expressed high levels of genes related to migration and invasion, including CTHRC1, TWIST1, POSTN, LOXL2, PDGFRB, and MMP14. Accordingly, in a transwell matrix invasion assay, both CD34−THY1+ and CD34+ populations showed enhanced migration and invasion (6).
CD34+ fibroblasts were more strongly associated with leukocyte recruitment. They expressed CXCL12, CCL2 and IL-6, and showed significantly higher secretion of these proteins in response to TNF-α exposure. They also recruited significantly higher numbers of leukocyte peripheral blood monocytes in a transwell leukocyte recruitment assay. Together, these data indicate that of the synovial fibroblasts studied, CD34+ subsets are likely to predominate in leukocyte recruitment to the inflamed synovium (6).
Taken together, this new work suggests that the CD34−THY1+ subset plays the strongest role in promoting synovial swelling and inflammation at both the molecular and clinical levels, followed by CD34+ subsets and lastly CD34−THY1− fibroblasts (6) (Table 1). There was no correlation between the proportion of CD34−THY1+ fibroblasts and disease duration, indicating that these fibroblast subsets are not a subsequent effect of chronic inflammation, and may play a causative role, an important data-driven hypothesis that now warrants further examination.
Synovial fibroblasts participate in devastating bone destruction which can occur when osteoclastogenic factor TNFSF11 [also known as receptor activator of NF-κB ligand (RANKL)] outcompetes RANK decoy receptor TNFRSF11B (7). The CD34−THY1+ subset expressed the highest ratio of TNFSF11 to TNFRSF11B, suggesting that this subset may predominantly direct bone resorption.
Interestingly, almost all synovial fibroblasts were positive for cadherin-11 (CDH11) (6), which is a known correlate of pathology (8,9). CDH11 also enhances synovial fibroblast migration and invasion, and is involved in the stimulation of synovial fibroblasts to release cytokines and chemokines (9,10). This new work from Mizoguchi et al. reveals new markers and functional clues towards manifestations of pathogenic behaviour within the broader CDH11 positive phenotype.
This important study by Mizoguchi et al. is a welcome step forward for the field. It presents a new roadmap to investigate fibroblast heterogeneity and function in the pathogenesis of RA, including the possibility of novel patient stratification options and strategies to evaluate treatment success. Their approach is a valuable resource to the immunological community and will assist in the identification of disease-associated fibroblast subsets in other human diseases. Future work to synthesise these and other synovial phenotypes identified at earlier stages of RA (4,5) will be important to identify key developmental relationships and pathways, as we strive to understand what drives the emergence of pathogenic synovial fibroblast subsets and how to clinically target them.
Acknowledgements
MH Abuwarwar is supported by the Saudi Arabian Cultural Mission, affiliate to the Royal Embassy of Saudi Arabia in Australia, and Ministry of Education, Saudi Arabia.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Gibofsky A. Overview of epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis. Am J Manag Care 2012;18:S295-302. [PubMed]
- Turner JD, Filer A. The role of the synovial fibroblast in rheumatoid arthritis pathogenesis. Curr Opin Rheumatol 2015;27:175-82. [Crossref] [PubMed]
- Lefevre S, Meier FMP, Neumann E, Muller-Ladner U. Role of synovial fibroblasts in rheumatoid arthritis. Curr Pharm Des 21:130-41. [Crossref] [PubMed]
- Filer A, Ward LSC, Kemble S, et al. Identification of a transitional fibroblast function in very early rheumatoid arthritis. Ann Rheum Dis 2017;76:2105-12. [Crossref] [PubMed]
- Karouzakis E, Raza K, Kolling C, et al. Analysis of early changes in DNA methylation in synovial fibroblasts of RA patients before diagnosis. Sci Rep 2018;8:7370. [Crossref] [PubMed]
- Mizoguchi F, Slowikowski K, Wei K, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 2018;9:789. [Crossref] [PubMed]
- Danks L, Komatsu N, Guerrini MM, et al. RANKL expressed on synovial fibroblasts is primarily responsible for bone erosions during joint inflammation. Ann Rheum Dis 2016;75:1187-95. [Crossref] [PubMed]
- Lee DM, Kiener HP, Agarwal SK, et al. Cadherin-11 in Synovial Lining Formation and Pathology in Arthritis. Science 2007;315:1006-10. [Crossref] [PubMed]
- Chang SK, Noss EH, Chen M, et al. Cadherin-11 regulates fibroblast inflammation. Proc Natl Acad Sci U S A 2011;108:8402-7. [Crossref] [PubMed]
- Noss EH, Chang SK, Watts GFM, et al. Modulation of matrix metalloproteinase production by rheumatoid arthritis synovial fibroblasts after cadherin 11 engagement. Arthritis Rheum 2011;63:3768-78. [Crossref] [PubMed]