
H3K27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR
Introduction
Lung cancer, with approximately 1.8 million new cases and more than 1 million related deaths each year, is the most common malignancy and has rapidly emerged as a primary cause of cancer-related death worldwide over the past several decades (1). Small cell lung cancer (SCLC) is the most aggressive type of lung cancer and accounts for approximately 15–18% of all lung cancer cases (2). The majority of SCLC patients are initially responsive to chemotherapy drugs, but they inevitably develop multidrug resistance within a year (3,4). Thus, a better understanding of the mechanisms underlying multidrug resistance is urgently needed for improving SCLC treatment.
HOX transcript antisense RNA (HOTAIR), a widely studied long non-coding RNA (lncRNA), is transcribed from the HOXC gene cluster, bridges polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1) and suppresses the transcription of HOXD genes by catalyzing H3 lysine 27 trimethylation (H3K27me3) and the spontaneous demethylation of H3K4 (5-7). Many researchers have also demonstrated that HOTAIR and enhancer of zeste homolog 2 (EZH2) interact with each other in different types of cancer, inducing gene silencing and influencing chromatin dynamics (7-11). Moreover, aberrant DNA methylation has been demonstrated to participate in the development of chemoresistance in various types of cancer, such as ovarian and lung cancer (12). In our previous study, HOTAIR knockdown decreased HOXA1 DNA methylation by reducing the expression of DNMT1 and DNMT3b in chemoresistant SCLC (13).
Histone methylation is a vital epigenetic phenomenon that participates in a diverse array of cellular processes, such as transcription control, RNA splicing and tumorigenesis (14-19). H3K27me3, which is catalyzed by EZH2 (20), is correlated with transcriptional repression, is essential for cancer chemoresistance and is a prognostic factor in lung cancer (21,22). Multiple studies have shown that HOTAIR couples with EZH2 to mediate both cell invasion and cell metastasis (23,24). However, whether HOTAIR affects HOXA1 DNA methylation in SCLC by regulating EZH2-mediated H3K27me3 remains largely unknown.
In this study, we found that EZH2 and H3K27me3 enrichment levels were markedly high in SCLC tissues and multidrug-resistant SCLC cells. Moreover, H3K27me3 was related to multidrug resistance. We found that HOTAIR promoted EZH2 and EZH2-mediated H3K27me3. H3K27me3 affected the methylation level of HOXA1. Furthermore, H3K27me3 acted as a feedback regulator of HOTAIR. Thus, these results suggest that H3K27me3 induces multidrug resistance in SCLC by affecting HOXA1 DNA methylation via the regulation of the lncRNA HOTAIR.
Methods
Cell lines and cell culture
The human SCLC cell lines NCI-H69 and NCI-H446 and the multidrug-resistant subline NCI-H69AR were purchased from the American Type Culture Collection (ATCC). Cells were cultured in RPMI 1640 containing 10% fetal bovine serum (Gibco, CA, USA) at 37 °C in a humidified incubator with 5% CO2.
Immunohistochemical (IHC) staining
Formalin-fixed, paraffin-embedded tissues were sectioned, deparaffinized and rehydrated gradually. Sample sections were incubated with primary antibodies directed against EZH2 (Cell Signaling Technology, #5426, USA) and H3K27me3 (Cell Signaling Technology, #9733, USA) at 4 °C overnight. Secondary antibodies were applied according to the manufacturer's protocol. The percentage of positive cells was calculated in five randomly selected high-power fields. The results were classified based on the total scores of the percentage of positively stained tumor cells and the staining intensity. The percentages of positively stained tumor cells were classified into four groups with scores ranging from 0 to 3 (<10%, 0; >10%, 1; >25%, 2; and >50%, 3). The staining intensities were scored as follows: no staining, 0; low, 1; medium, 2; and high, 3). A final IHC score was calculated by adding the two scores; a score >3 indicated positive expression, and a score ≤3 indicated negative expression.
Western blotting
Protein was extracted from cells with RIPA lysis buffer, and the protein concentrations were detected using the standard BCA method (BCA™ Protein Assay Kit, CoWin Biosciences, China). Subsequently, each protein sample was separated on 12% SDS-PAGE gels and transferred onto PVDF membranes (Millipore, Etten-Leur, Netherlands). After blocking with 5% nonfat milk for 2 h, the membranes were incubated with the primary antibodies used for IHC at 4 °C overnight. The membranes were then incubated with a horseradish peroxidase-labeled secondary antibody. Finally, the protein bands were visualized using chemiluminescence (ECL) after being washed again with TBST.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cell lines with TRIzol reagent (Invitrogen, NY, USA) according to the manufacturer’s instructions. RT-PCR assays were performed to detect HOTAIR expression using a Prime Script RT reagent kit (TIANGEN, Beijing, China) according to the manufacturer’s protocol. The primers used in our study were as follows: HOTAIR, (F) 5'-GGTAGAAAAAGCAACCACGAAGC-3' and (R) 5'-ACATAAACCTCTGTCTGTGAGTGCC-3'; GAPDH, (F) 5'-GAGTCAACGGATTTGGTCGT-3' and (R) 5'-CATGGGTGGAATCATATTGGA-3'. Data collection was performed with an ABI Illumina instrument. The results were normalized to the expression of GAPDH. The relative expression of A was analyzed using the 2−ΔΔCt method.
Bisulfite sequencing PCR (BSP)
Cell DNA was isolated, bisulfate modified, and detected as previously described (20). The primer sequences for PCR were as follows: (F) 5'-GT TTAGTTTTAGAATAGAGGAGGTGGTT-3' and (R) 5'-AAAATCAATAAACCAA AACTC-3'. The region of the D promoter is shown in Figure S1 in the supplementary material.
Flow cytometry
Cells were treated with chemotherapy drugs for 24 h and collected for cell apoptosis and cell cycle analyses. Cell apoptosis assays were performed using an Annexin V eFluor™ 450/eFluor™ 660 kit (eBioscience, San Diego, USA) according to the manufacturer’s protocol. For cell cycle analysis, cells were fixed in 70% ethanol at 4 °C overnight. Subsequently, the cells were incubated with RNase and stained with Fixable Viability Dye eFluor™ 660. Finally, CellQuest Pro software was used for apoptosis analyses, and ModFit LT software was used for cell cycle analyses.
Cell counting kit-8 (CCK-8)
Cells were seeded in 96-well plates at 7×103 cells per well. Following culture for 6 h, the cells were treated with chemotherapy drugs [cisplatin (CDDP; Shandong, China), etoposide (VP-16; Jiangsu, China) and adriamycin (ADM; Jiangsu, China)] for 24 h. The absorbance at 450 nm was measured after incubation with 10 µL of CCK-8 reagent (Dojindo, Kumamoto, Japan) for approximately 4 h. Cells without chemotherapy drug treatment were used to indicate 100% survival. The assay was conducted in six replicate wells for each sample, and three parallel experiments were performed.
Cell transfection
For stable transfection, H69 and H446 cells were infected with HOTAIR lentiviral particles (GenePharma, Shanghai, China) and control lentiviruses according to the manufacturer’s instructions. For HOTAIR knockdown, cells were transfected separately with two siRNAs (GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen, NY, USA). The transfection efficiencies were detected by qRT-PCR. The siRNAs used were as follows: siHOTAIR#1, 5'-CCCAUGGACUCAUAAACAATT-3' and 5'-UUGUUUAUGAGUCCAUGGGTT-3'; siHOTAIR#2, 5'-GC CUUUGGAAGCUCUUGAATT-3' and 5'-UUCAAGAGCUUCCAAAGGCTT-3'.
Statistical analysis
All experiments were performed in triplicate. The data are shown as the means ± SD, and the statistical analyses were carried out with SPSS 22.0 software. Student’s t-test was used to evaluate the difference between two means, and comparisons between more than two groups were calculated using one-way ANOVA. P<0.05 was considered statistically significant (*P<0.05, **P<0.01).
Results
H3K27me3 induced multidrug resistance in SCLC
To determine if H3K27me3 is associated with SCLC, we performed IHC to detect H3K27me3 expression in five SCLC tissue samples. As EZH2 was previously demonstrated to catalyze H3K27 methylation (25), we also examined the expression of EZH2. H3K27me3 and EZH2 were detected predominantly in the cell nucleus. EZH2 and H3K27me3 levels were markedly higher in SCLC cells than in normal lung alveolar epithelial cells (Figure 1A).
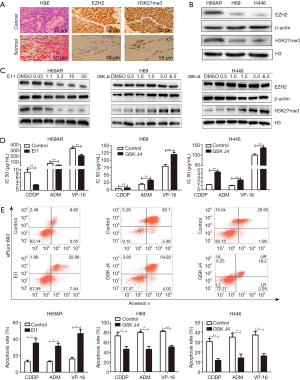
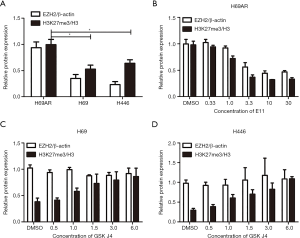
Then, we assessed H3K27me3 in H69AR/H69 and H446 cells with Western blotting assays and found that the H3K27me3 levels were increased in the multidrug-resistant cells compared with those in the parental cells (Figure 1B and Figure S2). We used highly selective inhibitors to alter the H3K27me3 levels. EI1, a selective inhibitor of EZH2, inhibits EZH2/PRC2 methyltransferase activity and decreases H3K27 methylation without changing other histone H3 methylation marks (25,26). GSK J4 is a selective inhibitor of two demethylases, JMJD3/UTX, the inhibition of which increases H3K27 methylation levels (27,28). EI1-treated cells (H69AR cells) exhibited decreased H3K27me3 upon EZH2 downregulation; in addition, GSK J4-treated cells (H69 and H446 cells) exhibited increased H3K27me3 with unchanged EZH2 levels (Figure 1C and Figure S2). CCK-8 assays indicated that the IC50 values of CDDP, ADM and VP-16 were significantly decreased in H69AR cells after treatment with EI1 and that the IC50 values significantly increased in H69 and H446 cells with GSK J4 treatment (Figure 1D). Flow cytometry showed that the apoptosis rates significantly increased in EI1-treated cells after anticancer drug treatment, whereas the apoptosis rates significantly decreased in GSK J4-treated cells (Figure 1E). Furthermore, cell cycle progression was not significantly changed (Figure S3). Thus, all these data indicate that H3K27me3 induces multidrug resistance and exerts antiapoptotic effects in SCLC.
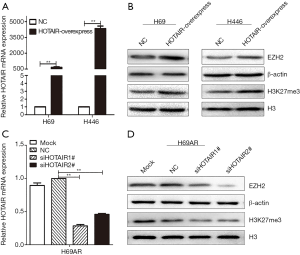
EZH2 and H3K27me3 were upregulated by HOTAIR
To determine whether HOTAIR plays a key role in EZH2 and H3K27 trimethylation, we examined the enrichment levels of EZH2 and H3K27me3 under HOTAIR interference. We previously found that HOTAIR mRNA levels were higher in H69AR cells than in H69 and H446 cells (13). We then used a lentivirus to overexpress HOTAIR by nearly 658-fold in H69 cells and 4340-fold in H446 cells (Figure 2A). HOTAIR overexpression significantly increased H3K27me3 and EZH2 levels in H69 and H446 cells according to Western blotting assays (Figure 2B). In contrast, HOTAIR expression was downregulated by as much as 70% in H69AR cells transfected with two different siRNAs (Figure 2C). Strikingly, silencing HOTAIR significantly reduced the levels of both EZH2 and H3K27me3 (Figure 2D). Collectively, these data suggest that EZH2 and H3K27me3 are upregulated by HOTAIR.
HOTAIR affected HOXA1 DNA methylation through H3K27me3
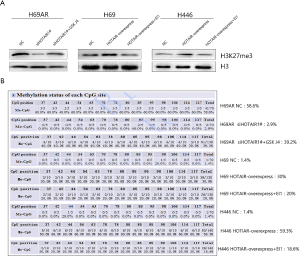
To explore whether HOTAIR affects HOXA1 DNA methylation by regulating H3K27me3, we used BSP to investigate the methylation status of the CpG island in the HOXA1 promoter. The analysis revealed that HOTAIR overexpression resulted in increased H3K27me3 levels and CpG island methylation levels in the HOXA1 promoter in H69 and H446 cells. In contrast, HOTAIR knockdown resulted in decreased H3K27me3 levels and CpG island methylation levels in the HOXA1 promoter in H69AR cells (Figure 3A). Next, we found that H3K27me3 levels decreased with EI1 treatment and that HOXA1 DNA methylation levels decreased consistently with H3K27me3 downregulation in HOTAIR-overexpressing cells (H69 and H446). Conversely, H3K27me3 levels increased with GSK J4 treatment and HOXA1 DNA methylation levels increased consistently with H3K27me3 upregulation in HOTAIR-knockdown cells (H69AR) (Figure 3B). These results indicate that H3K27me3 affects HOXA1 DNA methylation via HOTAIR regulation.
H3K27me3 acted as a negative feedback regulator of HOTAIR
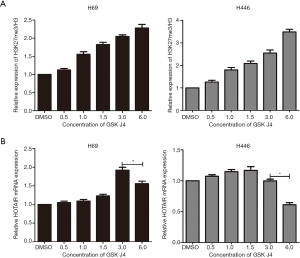
To further identify the association between H3K27me3 and HOTAIR, we investigated whether HOTAIR expression is dependent on H3K27me3 levels. H3K27me3 was dramatically increased in H69 and H446 cells treated with GSK J4 according to Western blotting assays (Figure 1C and Figure 4A). This increase occurred in a dose-dependent manner. However, after treatment with the same concentration of GSK J4, qRT-PCR indicated that HOTAIR expression levels constantly increased and reached a peak of 3.0 µM in H69 cells and of 1.5 µM in H446 cells and then decreased (Figure 4B). These results indicate that H3K27me3 acts as a negative feedback regulator of HOTAIR.
Discussion
SCLC has been designated a “recalcitrant” cancer due to its high lethality and the lack of substantial therapeutic progress made over the past years. Recent studies have revealed that the lncRNA HOTAIR acts as a vital mediator of the molecular mechanisms underlying cancer development and progression, including proliferation, metastasis and chemoresistance (13,23,29,30). Our previous studies showed that HOTAIR regulates HOXA1 DNA methylation by reducing DNMT1 and DNMT3b levels in chemoresistant SCLC. Moreover, HOTAIR couples with EZH2 to act as a pivotal mediator of cell invasion and metastasis. EZH2 possesses histone methyltransferase activity and methylates H3K27 (26,31). Recently, mounting evidence has suggested that H3K27me3 is correlated with chemoresistant cancer, including SCLC (21,22). To verify the hypothesis that H3K27me3 affects HOXA1 DNA methylation via HOTAIR regulation in SCLC chemoresistance, we initially found that EZH2 and H3K27me3 levels were markedly higher in SCLC tissues than in adjacent noncancerous tissues. Furthermore, we found that EZH2 and H3K27me3 were upregulated in multidrug-resistant SCLC cells. To further validate the biological functions of H3K27me3, we determined the effect of H3K27me3 on SCLC cell sensitivity to anticancer drugs. Our data indicated that H3K27me3 affects multidrug resistance by inhibiting apoptosis in SCLC. However, previous studies have shown that H3K27me3 suppresses proliferation via apoptosis and cell cycle arrest in cancer (32,33). This inconsistency may be attributed to the different types of cancer.
To further investigate whether EZH2 and H3K27me3 are regulated by HOTAIR, we successfully established two stable HOTAIR-overexpressing cell sublines and effectively suppressed HOTAIR expression using siRNAs. HOTAIR overexpression significantly increased EZH2 and H3K27me3 levels, whereas HOTAIR knockdown significantly reduced both EZH2 and H3K27me3 levels. Our results indicate that EZH2 and H3K27me3 are upregulated by HOTAIR.
CpG island methylation in the promoter may be a crucial mechanism regulating gene expression at the epigenetic level (34,35). Moreover, histone modifications alter DNA methylation (36,37). HOTAIR has been found to bind PRC2, mediate H3K27 trimethylation and silence the HOXD locus (38). To investigate whether HOTAIR affects HOXA1 DNA methylation through H3K27me3, we determined the correlation between the CpG island methylation status and H3K27me3 levels. HOXA1 DNA methylation levels decreased consistently with H3K27me3 downregulation and increased consistently with H3K27me3 upregulation, which validated our hypothesis. However, the molecular mechanisms involved in H3K27me3-induced HOXA1 DNA methylation need to be further explored in the future.
A previous study indicated that EZH2-mediated H3K27 methylation contributes to the repression of a subset of lncRNAs in embryonic stem cells (10). Our study provides evidence to support the hypothesis that H3K27me3 may repress HOTAIR expression. HOTAIR expression constantly increased and reached its peak level and then decreased dependent on increasing levels of H3K27me3. Additionally, H3K27me3 can bind the HOTAIR promoter region in human liver cancer stem cells (39). Therefore, all the results indicate that H3K27me3 suppresses HOTAIR, likely by binding to its promoter. To the best of our knowledge, this is the first study to reveal a negative feedback loop between H3K27me3 and HOTAIR in SCLC cells. This information may aid in understanding the pleiotropic functions of H3K27 methylation in the regulation of lncRNA expression in cancer progression. However, further experiments need to be performed to determine the mechanism by which H3K27me3 suppresses HOTAIR expression.
Conclusions
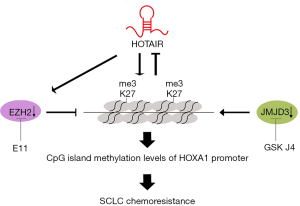
In conclusion, our study found that H3K27me3 induced multidrug resistance in SCLC by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR. Moreover, H3K27me3 modulated HOTAIR expression through a negative feedback mechanism (Figure 5). Therefore, our study aided in understanding the mechanism of SCLC chemoresistance and indicated that H3K27me3 may be a potential therapeutic target for treating chemoresistance in SCLC, but this treatment must be confirmed with additional clinical studies.
Acknowledgements
Funding: This work was supported by the Natural Science Foundation of Guangdong Province (2016A030310385) and the National Natural Science Foundation of China (81601986).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Planchard D, Le Pechoux C. Small cell lung cancer: new clinical recommendations and current status of biomarker assessment. Eur J Cancer 2011;47 Suppl 3:S272-83. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Kalemkerian GP, Schneider BJ. Advances in Small Cell Lung Cancer. Hematol Oncol Clin North Am 2017;31:143-56. [Crossref] [PubMed]
- Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012;31:4577-87. [Crossref] [PubMed]
- Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013;12:433-46. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long non-coding RNA as modular scaffold of histone modification complexes. Science 2010;329:689-93. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long non-coding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Wu SC, Kallin EM, Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res 2010;20:1109-16. [Crossref] [PubMed]
- Wu L, Murat P, Matak-Vinkovic D, et al. Binding interactions between long non-coding RNA HOTAIR and PRC2 proteins. Biochemistry 2013;52:9519-27. [Crossref] [PubMed]
- Teschendorff AE, Lee SH, Jones A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med 2015;7:108. [Crossref] [PubMed]
- Fang S, Gao H, Tong Y, et al. Long non-coding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest 2016;96:60-8. [Crossref] [PubMed]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41-5. [Crossref] [PubMed]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012;13:343-57. [Crossref] [PubMed]
- Kahn TG, Schwartz YB, Dellino GI, et al. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem 2006;281:29064-75. [Crossref] [PubMed]
- Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010;142:967-80. [Crossref] [PubMed]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 2010;10:457-69. [Crossref] [PubMed]
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol 2010;21:209-20. [Crossref] [PubMed]
- Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010;29:1681-90. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Avila-Moreno F, Armas-Lopez L, Alvarez-Moran AM, et al. Overexpression of MEOX2 and TWIST1 is associated with H3K27me3 levels and determines lung cancer chemoresistance and prognosis. PLoS One 2014;9:e114104. [Crossref] [PubMed]
- Wu Y, Zhang L, Zhang L, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 2015;46:2586-94. [Crossref] [PubMed]
- Zheng J, Xiao X, Wu C, et al. The role of long non-coding RNA HOTAIR in the progression and development of laryngeal squamous cell carcinoma interacting with EZH2. Acta Otolaryngol 2017;137:90-8. [Crossref] [PubMed]
- Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A 2012;109:21360-5. [Crossref] [PubMed]
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002;298:1039-43. [Crossref] [PubMed]
- Xiang Y, Zhu Z, Han G, et al. JMJD3 is a histone H3K27 demethylase. Cell Res 2007;17:850-7. [Crossref] [PubMed]
- Yan N, Xu L, Wu X, et al. GSKJ4, an H3K27me3 demethylase inhibitor, effectively suppresses the breast cancer stem cells. Exp Cell Res 2017;359:405-14. [Crossref] [PubMed]
- Sun L, Fang J. Writer meets eraser in HOTAIR. Acta Biochim Biophys Sin (Shanghai) 2011;43:1-3. [Crossref] [PubMed]
- Woo CJ, Kingston RE. HOTAIR lifts non-coding RNAs to new levels. Cell 2007;129:1257-9. [Crossref] [PubMed]
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 2010;35:323-32. [Crossref] [PubMed]
- Pawlyn C, Bright MD, Buros AF, et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer J 2017;7:e549. [Crossref] [PubMed]
- Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015;6:537-46. [PubMed]
- Kumegawa K, Maruyama R, Yamamoto E, et al. A genomic screen for long non-coding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci Rep 2016;6:26699. [Crossref] [PubMed]
- Merry CR, Forrest ME, Sabers JN, et al. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genet 2015;24:6240-53. [Crossref] [PubMed]
- Funata S, Matsusaka K, Yamanaka R, et al. Histone modification alteration coordinated with acquisition of promoter DNA methylation during Epstein-Barr virus infection. Oncotarget 2017;8:55265-79. [Crossref] [PubMed]
- Inoue A, Jiang L, Lu F, et al. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017;547:419-24. [Crossref] [PubMed]
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by non-coding RNAs. Cell 2007;129:1311-23. [Crossref] [PubMed]
- An J, Wu M, Xin X, et al. Inflammatory related gene IKKalpha, IKKbeta, IKKgamma cooperates to determine liver cancer stem cells progression by altering telomere via heterochromatin protein 1-HOTAIR axis. Oncotarget 2016;7:50131-49. [Crossref] [PubMed]



