
AMBRA1-mediated autophagy and apoptosis associated with an epithelial-mesenchymal transition in the development of cleft palate induced by all-trans retinoic acid
Introduction
Autophagy is a self-degradative process involved in the elimination of dysfunctional cellular components or damaged organelles (1-3) and acts as a pro-survival or pro-death mechanism during embryonic development, such as in palatogenesis. Apoptosis and epithelial-mesenchymal transition (EMT) are fundamental physiological processes, which are independent or interrelated events in normal development and in maintaining body homeostasis (1,2). Fusion of the palatal shelves, which involves EMT of the medial edge epithelium (MEE), is a crucial step in palate development. Interference with EMT of the MEE can be responsible for cleavage of the palate. Several lines of evidence suggest that apoptosis, migration of the cells toward the nasal and oral epithelium, and EMT is involved in the disappearance of the medial epithelial seam (MES) (4,5). For example, in mice, the palatal shelves grow into the midline, and palatal fusion occurs at embryonic gestation day 14.5 (E14.5); any imbalance of embryonic palatal mesenchyme cell proliferation and apoptosis can result in cleft palate (CP) formation (6-8).
Autophagy/beclin 1 regulator 1 (AMBRA1) is an important factor, at the crossroads between autophagy and apoptosis, that controls the reciprocal conversion between autophagy and apoptosis to decide cell death or survival (9-12). AMBRA1 regulates autophagy by promoting the formation of autophagosomes (13). Sun et al. have shown that functional defects of AMBRA1 in mouse embryos result in severe neural tube defects associated with autophagy and apoptosis (11). A previous study showed that lymphoid enhancer-binding factor 1 (LEF1) is involved in cleft lip and palate formation (14). LEF1-induced EMT may result in the disappearance of the palatal seam, along with strong MEE adherence and seam formation (15). The regulatory mechanisms of AMBRA1-mediated autophagy and apoptosis in palatogenesis, and how these mechanisms are associated with EMT in embryonic development and palatal shelf fusion, remain unclear. An increasing amount of research suggests that altered expression of long noncoding RNA (lncRNA) is associated with the genesis and progression of many human diseases (16,17). LncRNA dysregulation has been associated with various disorders, including CP (18). However, how specific lncRNAs affect the autophagy and apoptosis associated with EMT underlying palatogenesis, is not yet known.
All-trans retinoic acid is a metabolite of vitamin A and functions to support normal pattern formation during embryogenesis (19). Abnormally high concentrations of all-trans retinoic acid were reported to induce fetal malformations, including cleft palate, in both experimental animals and humans (20). In the present study, we used a CP model, established in C57BL/6J mice, to examine whether AMBRA1 can directly or indirectly mediate autophagy and apoptosis associated with EMT during palate shelf fusion.
Methods
Animals and treatment
C57BL/6J mice (weighing 20–28 g, from 8 to 10 weeks of age) were purchased from Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China). Females were mated with males overnight with similar weight and ages, and embryonic gestation day 0.5 (E0.5) was designated at 8 a.m. of the next day when a vaginal plug was found. Pregnant females were randomly divided into two groups. ATRA-treated groups were administered all-trans retinoic acid by gavage at E10.5. Control groups were given an equivalent volume of corn oil. Pregnant mice were sacrificed by cervical dislocation at E14.5. The fetuses were removed from the uterus, and the palatal shelves were harvested under sterile conditions with the stereomicroscope (SMZ 1500; Nikon Instruments, Melville, NY, USA). For this study, an ATRA-induced mouse CP model was used, and palatal shelf tissues were obtained and stored as reported previously (21,22). The animal study protocol was approved by the Laboratory Animal Ethical Committee of the Medical College of Shantou University (Shantou, China), and experiments were carried out by the animal care guidelines of the US National Institutes of Health.
Morphology and histology analysis
After palatal shelves were harvested, they were then immediately photographed using a Leica photographic system (M 10, Germany) for observation of the morphology of the palatal shelf. Next, the palatal shelves were fixed immediately in 4% paraformaldehyde/phosphate-buffered saline (PH 7.4) for 24 h. The tissues were processed for dehydration in alcohol gradient and embedded in paraffin. Serial sections (5 µm) were cut and stored at room temperature (20–25 °C). The sections were stained with hematoxylin and eosin for routine histology.
RNA extraction, library preparation, and RNA-seq
Total RNA was extracted from palatal shelf tissues of the ATRA-treated samples and control samples using a mirVana™ miRNA Isolation Kit (miRNA ISO lation Kit, Ambion, Carlsbad, CA, USA) following the manufacturer’s protocol. RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA Integrity Number (RIN) ≥7 were subjected to subsequent analysis. The libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina Inc., San Diego, CA, USA) according to the manufacturer’s instructions. The libraries were then sequenced on the Illumina sequencing platform by Shanghai Oebiotech Co. Ltd (Shanghai, China) (HiSeq™ 2500) and 125/150 bp paired-end reads were generated.
Sequence data processing and differential mRNA-lncRNA expression analyses
The raw data file obtained by high-throughput sequencing was stored in FASTQ (fq) file format. After adaptor sequences and low-quality sequences were removed from the original reads, the high-quality clean reads were mapped to the Mouse Genome (
LncRNA-mRNA correlation analysis
The network between lncRNA-mRNA was constructed based on the correlation analysis among differentially expressed lncRNAs and protein-coding genes. For each pair, the Pearson correlation was performed to assess the correlation. lncRNA-mRNA co-expressions with correlation coefficients of P<0.05 and an absolute value correlation coefficient of COR >0.7 was were considered statistically significant.
LncRNA-mRNA co-expression analysis
For each lncRNA, we calculated the Pearson correlation of its expression with the expression of each mRNA. For function prediction of lncRNAs, we firstly calculated co-expressed mRNAs for each differentially expressed lncRNA (28), and then performed Gene Ontology/Kyoto Encyclopedia of Genes and Genomes analyses (GO/KEGG) analyses for co-expressed mRNAs. The enriched functional terms were used to predict the biological role of the given lncRNA. The Pearson correlation coefficient was used to authenticate the co-expressed mRNA of the lncRNA. A hypergeometric distribution test was used to calculate the GO/KEGG terms to annotate the co-expressed mRNAs (29). The false discovery rate was calculated by a previously described method (30). LncRNA–mRNA co-expression with correlation coefficients of P<0.05 and an absolute value of the correlation coefficient COR >0.7 were considered potentially relevant.
LncRNA-TF-target gene network construction
LncRNA sequences were mapped to the genome in the NONCODE (V5) database (31). Jemboss software was used to examine the alignment of lncRNA and putative TF binding sequences (http://emboss.sourceforge.net/Jemboss) (32). The genome browser database was used to build the network describing the relationships between TFs and lncRNAs (https://genome.ucsc.edu) (33). Pearson correlation coefficients were used to authenticate the co-expressed TFs of the lncRNAs (P<0.05). A hypergeometric distribution test was used to calculate the GO/KEGG terms to annotate co-expressed TFs. The “TF-lncRNA” network was constructed using Cytoscape software (34).
According to the interactions of lncRNAs and target gene assemble of the TF/chromatin regulatory complex, the “lncRNA-TF-target gene” network was constructed. The degree of enrichment of the intersection was calculated by hypergeometric distributions, and the TFs that were significantly related to the lncRNAs were obtained, thus identifying the TF/chromatin regulatory factor that might be associated with lncRNAs. Using hypergeometric distributions, each lncRNA can form multiple lncRNA-TF pairs. Each “lncRNA-TF” pair is the result of multiple gene enrichment. Based on the P value distribution (low to high), the two-element relation graph used the regulatory relationship of the first 100 hits, and the three-relation network’s graph took the mapping relationship between the ten hits. The “lncRNA-TF-target gene” trans-regulatory network was constructed using Cytoscape software based on the “TF-lncRNA” network (34).
To further study the implication between autophagy and apoptosis of susceptibility genes and CP during palatal fusion, using the RNA-seq data and relevant bioinformatics analysis, we screened for genes that fulfilled the following conditions: (I) the co-expressed mRNA of the lncRNA analysis must meet the lncRNA-TF-target gene; (II) CP-related genes must be the central nodes of the lncRNA-TF-target gene network; (III) enrichment for signaling pathway of CP-related gene must be associated with palatal fusion. In this way, the “NONMMUT034790.2-LEF1-AMBRA1” trans-regulatory network was determined.
qPCR validation
qPCR was conducted in six individual samples to validate the RNA-seq data (triplicates). The qPCR primers used in this study are listed in Table 1. The mRNA expression levels relative to lncRNA were analyzed as described in a previous study, and the 2−ΔΔCt method was used to calculate the level of gene expression relative to the expression of β-actin as an internal control (35).

Full table
Statistical analysis
PCR data were analyzed using Student’s t-test to compare the means between samples from control and ATRA-treated mice. A hypergeometric distribution test was used to calculate the GO/KEGG terms to annotate co-expressed TFs. LncRNA-mRNA co-expression levels were analyzed using the Pearson correlation. A P value <0.05 and log2FC >1 were considered statistically significant.
Results
Morphology and histology of embryonic palate shelf tissue
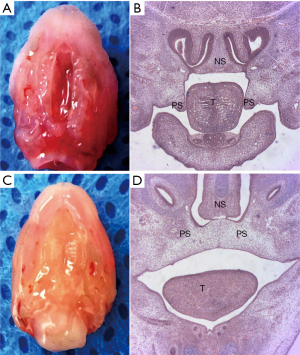
In the palate shelf tissue and histological sections of control E14.5 embryos, it was observed that the palatal shelf had already contacted the midline and fused through by formation of the midline epithelial seam (MES) in the mid-anterior region, whereas in palate shelf tissue and histological sections from ATRA-treated embryos, it was observed that the palatal shelf had separated without fusion (Figure 1).
Overview of lncRNA-mRNA co-expression profiles
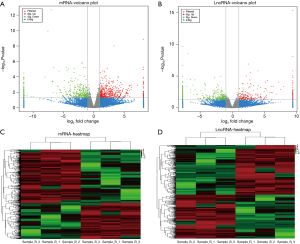
To determine the mRNA-lncRNA co-expression profiles that are differentially expressed in the palate between control and ATRA-treated mice at E14.5, we performed a lncRNA-mRNA-seq data co-expression analysis of ATRA-treated and control tissues of embryonic palatal shelves. A total of 703 differentially expressed mRNAs and 583 differentially expressed lncRNAs were identified between ATRA-treated vs. control groups. First, the expression level for each mRNA and lncRNA was included in a volcano plot (Figure 2A,B). Next, the differentially expressed mRNAs and lncRNAs were also used to perform a heat map-based unsupervised hierarchical clustering analysis (Figure 2C,D). Among the differential mRNA-lncRNA co-expressions, the NONMMUT034790.2 (P=0.024, log2FC =−1.6)/LEF1 (P=0.0009, log2FC =−1.5)/AMBRA1 (P=0.0386, log2FC =−1.3) was downregulated (http://atm.amegroups.com/public/system/atm/supp-atm.2019.02.22-1.pdf; http://atm.amegroups.com/public/system/atm/supp-atm.2019.02.22-2.pdf).
LncRNA-mRNA correlation analysis
To further determine whether NONMMUT034790.2 was correlated with LEF1/AMBRA1 during palatal fusion, correlation analysis of lncRNA-mRNA was performed using the results of the lncRNA-mRNA-seq data. A total of 584 lncRNAs relative to 703 mRNAs were exhibited (P<0.05 and COR >0.7). Among the lncRNA-mRNA correlations, NONMMUT034790.2 relative to AMBRA1 gave values of P=0.033 and COR =0.85, while NONMMUT034790.2 relative to LEF1 resulted in P=0.037 and COR =0.84 (http://atm.amegroups.com/public/system/atm/supp-atm.2019.02.22-3.pdf).
LncRNA-mRNA co-expression analysis and function annotation
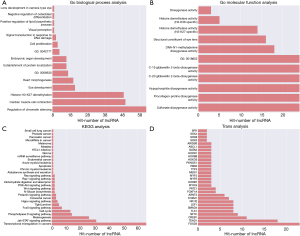
The lncRNA-mRNA co-expression function annotation was used in GO/KEGG analysis by selecting the reliability prediction terms (P<0.05, fold enrichment >2). Two hundred prediction relationships with the highest predictive credibility according to the P value ranking were selected to be representative of GO terms for biological processes or molecular functions (Figure 3A,B). The GO analyses showed that expression-linked lncRNA-mRNA genes were associated with an “intrinsic apoptotic signaling pathway in response to DNA damage by the p53 class mediator”, “embryonic organ development”, and an “intrinsic apoptotic signaling pathway in response to DNA damage”. Moreover, Figure 3C indicates that expression-linked lncRNA-mRNA gene KEGG pathways were involved in “apoptosis”, the “PI3K-Akt signaling pathway”, the “Wnt signaling pathway”, and the “Hippo signaling pathway”.
We then specifically analyzed and identified the AMBRA1 with a biological process. AMBRA1 was involved in “positive regulation of mitophagy”, “regulation of autophagosome assembly”, and “positive regulation of phosphatidylinositol 3-kinase activity”. NONMMUT034790.2 was involved in “positive regulation of mitophagy”, and “regulation of autophagosome assembly”. LEF1 was involved in “positive regulation of the cell cycle process”, the “apoptotic process involved in morphogenesis,” and “positive regulation of the epithelial to mesenchymal transition” (Table 2) (P<0.05, fold enrichment >2).

Full table
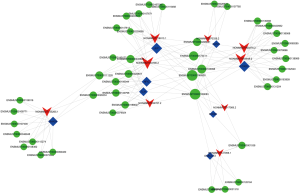
Constructed “lncRNA-TF-target gene” trans-regulatory network
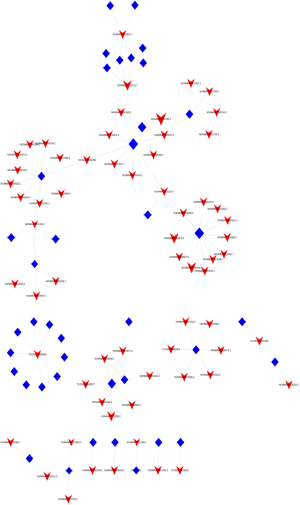
Trans-regulatory mechanisms involve TF-mediated chromatin regulation and transcription. Based on our lncRNA-mRNA co-expression analysis, a “lncRNA-TF” regulatory network was constructed using Cytoscape software. “NONMMUT034790.2”-LEF1 was located in the center of the network map between the ATRA-treated and control specimens (Figure S1). According to the results of the “lncRNA-TF” regulatory network analysis, we then constructed a “lncRNA-TF-target gene” trans-regulatory network using Cytoscape software. NONMMUT034790.2–LEF1-AMBRA1 was located in the center of the network map between the ATRA-treated and control specimens (Figures 3D,4).
qPCR validation
qPCR was performed to further validate the differential lncRNA-mRNA co-expression for NONMMUT034790.2-LEF1-AMBRA1. We found that the expression levels of NONMMUT034790.2 (P=4E-07), LEF1 (P=5E-06), and AMBRA1 (P=7E-05) mRNA were significantly downregulated in the samples from ATRA-treated mice compared to the control samples (Figure 5). The RNA-seq data of NONMMUT034790.2 (P=0.024, log2FC =−1.6)/LEF1 (P=0.0009, log2FC =−1.5)/AMBRA1 (P=0.0386, log2FC =−1.3) were downregulated. The qPCR results are well matched to the RNA-seq data.

Discussion
In this study, we predicted whether abnormal AMBRA1 has potential effects on embryonic mouse palate shelf fusion. Functional annotation showed that AMBRA1-mediated autophagy and apoptosis are associated with EMT by a “NONMMUT034790.2-LEF1-AMBRA1” trans-regulatory network during palatal fusion in ATRA-induced mouse CP. Data obtained from the mRNA-lncRNA co-expression analysis for AMBRA1, a key protein in autophagy regulation, identified “NONMMUT034790.2-LEF1-AMBRA1” as a trans-regulatory network for AMBRA1 regulation. This trans-regulatory mechanism of “NONMMUT034790.2-LEF1-AMBRA1” in palatogenesis, following ATRA-induced CP formation, has been supported in four ways: (I) the results of morphology and histology (Figure 1); (II) mRNA expression level changes of the “NONMMUT034790.2-LEF1-AMBRA1” co-expression profiles being downregulated in ATRA-treated vs. control embryonic palatal shelf tissues; (III) function annotation showing “NONMMUT034790.2-LEF1-AMBRA1” being significantly enriched in important biological processes related to autophagy and apoptosis associated with EMT (Figure 3); (IV) qPCR results being consistent with RNA-seq data.
CP is one of the most frequent congenital disabilities with a complex genetic and environmental etiology. However, the function and regulation of AMBRA1-mediated autophagy and apoptosis associated with EMT have rarely been reported. Neural crest cells that form the facial primordia originate on the dorsal neural tube (36). The palatal shelves consist mainly of cranial neural crest-derived mesenchymal cells covered by a simple embryonic epithelium (37). AMBRA1 is primarily expressed in the neural plate of the central nervous system during mouse embryogenesis (38). AMBRA1-deficiency in mouse embryos results in severe neural tube defects associated with autophagy and apoptosis (11). LEF1, as a transcript factor, appears to be required for the induction of genes responsible for periderm transition into mesenchymal tissue and subsequent palate formation (22). LEF1 promotes palatal EMT, and TGF-β3 stimulates LEF1 mRNA synthesis in MEE cells. AMBRA1 has been shown to promote autophagosome formation and further regulate autophagy (21). However, the way by which AMBRA1 triggers EMT is still unknown. AMBRA1-mediated autophagy promotes the induction of EMT, and activation of TGF-β/SMAD3-dependent signaling plays a key role in regulating autophagy-induced EMT (39). Conversely, decreased AMBRA1 expression can repress EMT (40), possibly implying a role for autophagy in the palatogenesis of CP. The AMBRA1-mediated induction of autophagy and the inhibition of apoptosis associated with EMT might be involved in palatal fusion (13).
In our study, we demonstrate that a decreased expression of AMBRA1 might inhibit EMT and promote palatal mesenchymal cell apoptosis, and this mechanism may be achieved through a “NONMMUT034790.2-LEF1-AMBRA1” trans-model. This is the first time that involvement of AMBRA1 is implicated in ATRA-induced CP, suggesting that AMBRA1 may be a potential CP therapeutic target. Also, we identified the reduced expression of LEF1, NONMMUT034790.2, and AMBRA1 in CP mice, supporting the notion that AMBRA1-mediated autophagy and apoptosis is associated with EMT through a trans-regulatory mechanism of “lncRNA-TF-target genes” that prevent CP during palatal fusion.
However, our current study is preliminary, and further research is needed to uncover how AMBRA1-mediated autophagy and apoptosis are associated with EMT results in CP formation. Our sample size was relatively small, and the palatal shelves were directly obtained from embryonic mouse tissues, which could be mixed with other tissues. Although a correlation of “NONMMUT034790.2-LEF1-AMBRA1” during palatal fusion in ATRA-induced CP was identified in our current study, the underlying mechanisms of how “NONMMUT034790.2-LEF1-AMBRA1” affects palatal fusion remain to be identified. We expect to integrate AMBRA1 into the trans-regulatory network, which will help to elucidate further the pathogenesis of AMBRA1-mediated autophagy and apoptosis associated with EMT by ATRA-induced CP.
Conclusions
In summary, our results reveal that AMBRA1 might be a novel epigenetic biomarker in palatogenesis and imply that AMBRA1-mediated autophagy and apoptosis associated with EMT by a “NONMMUT034790.2-LEF1-AMBRA1” trans-regulatory network might be an important mechanism underlying dysfunctional palatal fusion. These results lay the foundation for early clinical intervention, genetic diagnosis, and treatment of CP in the future. We will attempt to further clarify the relationship between “NONMMUT034790.2-LEF1-AMBRA1” and CP in the autophagic machinery in future studies.
Acknowledgements
Funding: This work was supported by the grants from the Natural Science Foundation of Guangdong Province (#2015A030313431).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The animal study protocol was approved by the Laboratory Animal Ethical Committee of Medical College of Shantou University (Shantou, China), and experiments were carried out by the animal care guidelines of the US National Institutes of Health.
References
- Capizzi M, Strappazzon F, Cianfanelli V, et al. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy 2017;13:554-66. [Crossref] [PubMed]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004;6:463-77. [Crossref] [PubMed]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest 2005;115:2679-88. [Crossref] [PubMed]
- Ke FFS, Vanyai HK, Cowan AD, et al. Embryogenesis and Adult Life in the Absence of Intrinsic Apoptosis Effectors BAX, BAK, and BOK. Cell 2018;173:1217-30.e17. [Crossref] [PubMed]
- Song J.. EMT or apoptosis: a decision for TGF-beta. Cell Res. 2007;17:289-90. [Crossref] [PubMed]
- Carette MJ, Ferguson MW. The fate of medial edge epithelial cells during palatal fusion in vitro: an analysis by DiI labelling and confocal microscopy. Development 1992;114:379-88. [PubMed]
- Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 1990;181:195-213. [Crossref] [PubMed]
- Nawshad A.. Palatal seam disintegration: to die or not to die? that is no longer the question. Dev Dyn 2008;237:2643-56. [Crossref] [PubMed]
- Fimia GM, Corazzari M, Antonioli M, et al. Ambra1 at the crossroad between autophagy and cell death. Oncogene 2013;32:3311-8. [Crossref] [PubMed]
- Rice DP. Craniofacial anomalies: from development to molecular pathogenesis. Curr Mol Med 2005;5:699-722. [Crossref] [PubMed]
- Sun WL. Ambra1 in autophagy and apoptosis: Implications for cell survival and chemotherapy resistance. Oncol Lett 2016;12:367-74. [Crossref] [PubMed]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131-42. [Crossref] [PubMed]
- Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007;447:1121-5. [PubMed]
- Gu W, Wan D, Qian Q, et al. Ambra1 is an essential regulator of autophagy and apoptosis in SW620 cells: pro-survival role of Ambra1. PLoS One 2014;9:e90151. [Crossref] [PubMed]
- Liu J, Chen Z, Guo J, et al. Ambra1 induces autophagy and desensitizes human prostate cancer cells to cisplatin. Biosci Rep 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Martinelli M, Carinci F, Morselli PG, et al. Evidence of LEF1 fetal-maternal interaction in cleft lip with or without cleft palate in a consistent Italian sample study. Int J Immunopathol Pharmacol 2011;24:15-9. [Crossref] [PubMed]
- Nawshad A, LaGamba D, Hay ED. Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT). Arch Oral Biol 2004;49:675-89. [Crossref] [PubMed]
- Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010;143:46-58. [Crossref] [PubMed]
- Ackermans MM, Zhou H, Carels CE, et al. Vitamin A and clefting: putative biological mechanisms. Nutr Rev 2011;69:613-24. [Crossref] [PubMed]
- Cuervo R, Valencia C, Chandraratna RA, et al. Programmed cell death is required for palate shelf fusion and is regulated by retinoic acid. Dev Biol 2002;245:145-56. [Crossref] [PubMed]
- Qin F, Shen Z, Peng L, et al. Metabolic characterization of all-trans-retinoic acid (ATRA)-induced craniofacial development of murine embryos using in vivo proton magnetic resonance spectroscopy. PLoS One 2014;9:e96010. [Crossref] [PubMed]
- Shu X, Shu S, Cheng H, et al. Genome-Wide DNA Methylation Analysis During Palatal Fusion Reveals the Potential Mechanism of Enhancer Methylation Regulating Epithelial Mesenchyme Transformation. DNA Cell Biol 2018;37:560-73. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511-5. [Crossref] [PubMed]
- Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621-8. [Crossref] [PubMed]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357-9. [Crossref] [PubMed]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 2013;10:71-3. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Fury W, Batliwalla F, Gregersen PK, et al. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc 2006;1:5531-4. [Crossref] [PubMed]
- Storey JD. A Direct Approach to False Discovery Rates. J R Stat Soc Series B Stat Methodol 2002;64:479-98. [Crossref]
- Fang S, Zhang L, Guo J, et al. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res 2018;46:D308-14. [Crossref] [PubMed]
- Carver TJ, Mullan LJ. JAE: Jemboss Alignment Editor. Appl Bioinformatics 2005;4:151-4. [Crossref] [PubMed]
- Casper J, Zweig AS, Villarreal C, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 2018;46:D762-9. [PubMed]
- Kohl M, Wiese S, Warscheid B.. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Marcucio R, Hallgrimsson B, Young NM. Facial Morphogenesis: Physical and Molecular Interactions Between the Brain and the Face. Curr Top Dev Biol 2015;115:299-320. [Crossref] [PubMed]
- Lan Y, Xu J, Jiang R.. Cellular and Molecular Mechanisms of Palatogenesis. Curr Top Dev Biol 2015;115:59-84. [Crossref] [PubMed]
- Benato F, Skobo T, Gioacchini G, et al. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy 2013;9:476-95. [Crossref] [PubMed]
- Scapoli L, Martinelli M, Arlotti M, et al. Genes causing clefting syndromes as candidates for non-syndromic cleft lip with or without cleft palate: a family-based association study.%A Scapoli L. Eur J Oral Sci 2008;116:507-11. [Crossref] [PubMed]
- Strappazzon F, Vietri-Rudan M, Campello S, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J 2011;30:1195-208. [Crossref] [PubMed]


