
Liquid biopsy for lung cancers: an update on recent developments
Introduction
Lung cancer is the most common cancer worldwide, accounting for 2.1 million new cases and 1.8 million deaths in 2018 (1). More than half of people with lung cancer die within one year of being diagnosed (2). When cancer is detected at a localized stage (I–II), the five-year survival rate is about 56 percent (3). However, only 16 percent of lung cancer cases are diagnosed at an early stage. For advanced/metastatic tumors (stage IV), the five-year survival rate is only 5 percent (4). Therefore, it is imperative to identify diagnostic methods for early detection of lung cancer, enabling a timely treatment plan while potentially reducing healthcare costs.
Early detection could be pursued using different approaches, such as diagnostic imaging techniques in high risk individuals, including low-dose computed tomography (LDCT). In the National Lung Screening Trial (NLST), LDCT showed a 20% decreased reduction in mortality from lung cancer compared to chest radiograph (5). More recently, the NELSON trial confirmed the value of LDCT screening in high-risk for lung cancer individuals. Overall, the LDCT scanning decreased mortality by 26% in high-risk men and 61% in high-risk women over a 10-year period (6). Nevertheless, LDCT has certain limitations when used as the only screening tool; high-false positive rate, over diagnosis, and increased radiation exposure. The detection of presumed early stage lung malignancies still requires tissue diagnosis. As technology continues to evolve in the field of interventional pulmonology, acquisition of tissue for diagnosis of lung lesions has become faster, safer and more precise (7). This field is rapidly growing and is not limited to diagnosis, but is also expanding to staging and treatment of lung cancer. An even less invasive modality that could be used as an alternative or complementary to both imaging and minimally invasive tissue acquisition would certainly be appealing. It would have the advantage of providing enough information for ‘individualized’ cancer therapy (molecular analysis). In this context, liquid biopsy or a blood specimen analysis, with assessment of various circulating tumor biomarkers has become a viable surrogate and is currently the subject of extensive research worldwide (8).
In this article, we will review the current molecular biomarkers available for liquid lung biopsy, their role in early detection, screening and prognostication of lung cancer and describe their use in clinical practice.
The concept of liquid lung biopsy and overview of its use in lung cancer
Unlike traditional biopsy, which usually involves either a surgical approach or a minimally invasive intervention to collect tumor tissue; liquid biopsy can easily be retrieved from plasma or serum. The role of liquid biopsy is mainly to determine actionable genomic alterations in different cancers that will eventually guide therapy and help assess response to targeted treatments. These liquid samples can be processed by different methods including, real time polymerase chain reaction-based methods (PCR or RT-PCR), droplet-digital polymerase chain reaction (ddPCR), Beads, Emulsion, Amplification and Magnetics (BEAMing), next generation sequencing (NGS) and other methods. To date, different circulating cancer biomarkers are being studied for liquid biopsy including: circulating tumor DNA (ctDNA), exosomes, tumor-associated antigens (TAAs), tumor educated platelets (TEP), tumor-associated autoantibodies (TAAbs), circulating tumor cells (CTCs), micro RNA among others (Table 1).
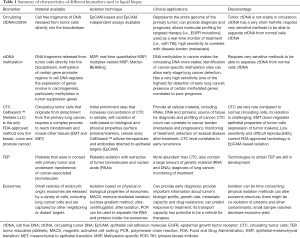
Full table
Currently, liquid biopsy is mainly utilized as an auxiliary tool in lung cancer diagnosis when a LDCT detects an abnormality. It also has a potential to be an integral part of lung cancer screening, early diagnosis, monitoring treatment response and prognostication.
The TRACERx study showed that ctDNA could be detected up to 6–12 months prior to cancer diagnosis using imaging, because genomic and chemical abnormalities related to cancer precede the appearance of detectable abnormalities in chest imaging (9). This could potentially eliminate or significantly reduce risk of radiation exposure by doing multiple follow up imaging for stable lung lesions and also reduce the complications related to high false positive rates seen in LDCT screening.
A plasma sample can potentially be used to analyze several biomarkers, like tumor cells or fragments of DNA from tumor cells and germline mutations; the information can be used towards a better characterization of tumor and with added advantage of being easily reproducible (blood draw) and with a safer profile. Shukuya et al. reported seven lung cancer cases in which they identified germline mutations using NGS that could be responsible for the development of lung cancer, and that identifying such germline mutations can have important implications when predicting cancer risk in other family members (10).
Positive liquid biopsy specimen for tumor driver mutation in otherwise healthy individual requires further workup for detection of clinical disease in the form of some imaging technique. In this context, another possibility is the chance of clonal hematopoiesis. Clonal hematopoiesis refers to the development of mutations in peripheral blood cells (PBC) that are not otherwise identified in matched specimens of cell-free DNA (cfDNA) and tumor tissue (11). This poses another challenge when interpreting cell-free ctDNA test results. Liquid biopsy should be complementary to other diagnostic tests.
Quantitative analysis of tumor ctDNA also correlates with disease burden and can be a useful tool for monitoring. Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) is a tool that can detect several genetic alterations in non-small cell lung carcinoma (NSCLC) patient in different stages. In 17 NCSLC patients, tumor ctDNA has been detected in 100% in stage II–IV patients and 50% in stage I patients. There is also high correlation between ctDNA level and tumor volume, which helps with the assessment of treatment response much earlier than radiographic changes (12). It is shown that ctDNA has a half-life of approximately 2 hours which allows evaluation of tumor changes in relatively short period of time (13).
Another important clinical application of liquid biopsy is monitoring for residual disease after surgery or chemotherapy. Current management is mainly based on TNM staging system but ctDNA can have important role to make decision to start adjuvant chemotherapy after curative intent surgery (14).
In the era of targeted or individualized cancer therapy with novel immuno chemotherapeutic agents, monitoring of emergent molecular resistance takes paramount importance. Liquid biopsy is also useful after incomplete response or recurrence of cancer, particularly to check for molecular resistance. It can also eliminate the necessity of doing repeat tissue biopsy for rechecking for genetic mutations (15). In 2016, the U.S. Food and Drug Administration (FDA) approved ctDNA as the first liquid biopsy test for NSCLC patients to check for EGFR-targeted therapy (16). In European Union there is established European consortium by European Liquid Biopsy Academy (ELBA) to investigate liquid biopsy for early detection of NSCLC using ctDNA, CTCs, exosomes, and TEPs bio sources (17).
There is data regarding the use of liquid biopsy in other body fluids (i.e., bronchial washings, pleural fluid, etc.). However, for the purpose of this review, we will focus on liquid biopsy performed in blood and/or plasma specimens only.
Biomarkers used in liquid lung biopsies
Circulating cfDNA and ctDNA
There are conditions in which cell free DNA is released in the bloodstream for instance in inflammatory states or pregnancy by mechanisms not yet fully understood (18). The concentration of free plasma DNA has been found to be higher in cancer patients compared to healthy subjects, difference driven mainly by the DNA derived from tumor cells (ctDNA) (19). It is not clear how ctDNA is released into the bloodstream, however, prior studies have suggested its source might be cell lysis from necrosis and/or apoptosis (20). It is tumor specific and provides molecular clues about fragmented DNAs of tumor cells and their specific mutations (21). In one of the initial studies, Sozzi et al. obtained the blood (7.5 mL sample) from one hundred patients with treatment-naïve NSCLC and matched it to appropriate controls. The concentration of plasma DNA in patients with lung cancer was close to eight times higher than the one seen in matched controls; higher levels of DNA in plasma also identified high-risk individuals (22).
ctDNA is usually found highly fragmented and at times mixed with non-tumor DNA. Discrimination of ctDNA from cfDNA deriving from normal cells is possible using ultrasensitive analytical assays (18). The presence of specific genomic alterations in ctDNA, including mutations in oncogenes or tumor suppressor genes, gene amplifications or epigenetic changes, are typically found in the cancer cell genome. The amount of ctDNA correlated with tumor burden, tumor response, and survival outcome.
Novel, highly sensitive blood-based assays to test cfDNA and ctDNA at very low concentrations for most genomic abnormalities have been developed. Studies have shown elevated concentrations of these in patients with early stage lung cancer, patients with recurrent disease and metastatic disease as compared to normal subjects (23). This makes it an attractive tool for prognosis, treatment response, surveillance of residual disease and progression and screening of high-risk patients. In this review, we will make efforts to refer to only ctDNA as the main biomarker used in different test platforms.
Methylated ctDNA
Recently identification of methylation of the ctDNA has emerged as a potential tool for diagnosis and screening. Methylation in specific tumor suppressor genes was more frequently observed in patients diagnosed with lung cancer than those with nonmalignant diseases. Hypermethylation has been shown to contribute to carcinogenesis and one of the main advantages of DNA methylation alterations, compared to other potential diagnostic biomarkers, is that they are remarkably stable, and generally occur early during carcinogenesis. Methylation in specific tumor suppressor genes, like Methyl Guanyl Methyl Transferase (MGMT), CDKN2A, Ras association domain-containing protein 1 (RASSF1A), Death-associated protein kinase 1 (DAPK) and Retinoic acid receptor beta (RAR-β), was more frequently observed in patients diagnosed with lung cancer than those with nonmalignant diseases. One report showed hypermethylation of some of the tumor suppressor genes was associated with poor prognosis (22).
The presence in plasma of methylated SHOX2 gene (a member of the homeobox family of genes that encode DNA binding transcription factors), has emerged as a sensitive and specific biomarker for lung cancer in different reports (24). SHOX2 gene hypermethylation was specifically associated with SCLC and squamous cell carcinoma. Recently it has been used as a marker in various other body samples of the patient like sputum, bronchoalveolar lavage, CSF, etc. (25). In one study SHOX2 methylation allowed to distinguish between malignant and benign lung diseases with a sensitivity of 68% and a high specificity of 95% based on testing from bronchial fluids. Epigenomics assays detects the relative amount of methylated SHOX2 gene fragments in a background of normal DNA in a real-time PCR assay based on TaqMan technology from bronchial fluid specimens. This test has been successfully validated as a sensitive and reliable lung cancer diagnostic tool from bronchial lavage specimens and can be used alongside other diagnostic procedures (18).
CTCs in lung cancer
CTCs were first described by Thomas Ashworth in 1869. He was able to isolate cells from the peripheral blood; they looked a lot like the cells from the primary cancer. CTC originate from detachment from a primary cancer. They have also been described to play a pivotal role in the metastatic spread of cancer (26).
It was not until more recently, that CTCs became a target of multiple studies in order to be used not only for diagnosis but also prognosis and to monitor treatment of cancer (27).
Initial local excessive cell proliferation and angiogenesis is a hallmark of cancer, however in order for a primary cancer to invade and metastasize into distant tissues, tumor cells need to first be able to gain certain properties. Epithelial tumor cells typically undergo a complex process known as epithelial-mesenchymal transition (EMT), where a cascade of signals enable these cells to enter the circulation and reach a remote target, where they will later undergo a reverse process called mesenchymal to epithelial transition (MET). Here, these cells will switch again from a mesenchymal into an epithelial phenotype, allowing further proliferation and formation of metastases (28).
Historically, isolation of CTC has been shown to be relevant for diagnosis and prognosis of cancers such as breast (29), prostate (30) and colon (31). It has the advantage of providing enough material (proteins, RNA and DNA) for molecular characterization and other analysis such as DNA studies. Presence of these CTCs is also considered a predicting factor for disease progression and clinical prognosis of patients with lung cancer (32).
Hannsen et al. studied a sample of patient with and without metastatic disease to correlate the positivity rate for CTC with the presence of metastases. They used a CTC kit intended for the enumeration of CTCs as well as multiplex RT-PCR (mRT-PCR) to look for expression of Ep-CAM and other genes (PIK3CA, AKT2, TWIST and ALDH1) in CTCs. They found a greater expression of these genes in the blood of patients with chemo-naïve metastatic disease and with progressive disease compared to those with non-metastatic lung cancer (M0). These findings were statistically significant; they were able to correlate the presence of CTC with disease burden, including metastases and progression (33). They also showed how the inclusion of other non-EpCAM-dependent methods increased the sensitivity to detect CTC.
NSCLC tends to recur locally or metastasize even after complete resection of the primary tumor in up to 45% of the cases. CTCs are suspected to be responsible of this. Despite the known aggressiveness and invasiveness of NSCLC, isolation of CTC has been challenging in this particular cancer. The initial EMT has the disadvantage that it down-regulates the epithelial properties of tumor cells, including the expression of epithelial tumor markers, which can difficult the detection of these markers by the existing techniques. The CellSearch® system (Janssen Diagnostics, LLC, Raritan, USA) is the most known U.S. FDA-approved CTC detection technology. It is able to isolate CTC with the use of anti-EpCAM ferromagnetic microbeads. However, it still lacks 100% sensitivity, maybe in part because of this downregulation of EpCAM during EMT (34).
EMT also causes increased expression of other mesenchymal markers in CTCs, such as N-cadherin, vimentin and transcriptional factors Zeb-1 and TWIST (35). Mahmood et al. showed a significant correlation between the expression of N-cadherin and vimentin and advanced tumor stage (36). Other authors that studied CTC in patients with prostate and breast cancers have also described that increased expression of N-cadherin and vimentin in CTC are associated with advanced disease.
As mentioned before in this review, use of ctDNA for detection of EGFR mutations is now a widely used tool, however CTC assays can enable EGFR detection as well, becoming a complementary test to ctDNA. Studies done on detection of EGFR mutations in CTC have clearly described how this can facilitate monitoring a patient’s response to targeted therapy and identify patterns of drug resistance (37). Other biomarkers have been included into the potential utility of CTC detection. Programmed death ligand-1 (PD-L1) expression on CTC has been reported in some studies; however, authors have not been able to describe a clear relationship between its expression and cancer stratification and prognosis in patients with NSCLC.
Clinical significance of CTC enumeration is yet to be established. Besides CellSearch® system, at this time the only existing FDA-approved EpCAM-dependent CTC detection method, other technologies able to detect different cellular and nuclear biomarkers have shown to have potential use for detection of early and minimal residual disease in NSCLC. It has been established that a high CTC count correlates with worse prognosis in cancers that have been studied, however, standardization in the use of CTC detection techniques and its count threshold is still required to assess its real clinical relevance as a liquid/blood-based biomarker. The ability to isolate enough CTC in the blood of NSCLC patients, identify its various biomarkers and obtain sequential sampling during treatment can potentially allow earlier detection of the lung cancer and will also provide real-time surveillance of cancer progression, monitoring of treatment response, prognosis and outcome.
Exosomes
Exosomes are very small cellular membrane-derived vesicles that measure between 30–100 nm. They are formed by endocytosis and are secreted by all types of cells (38). They can, therefore, be found in all types of body fluids, including blood (plasma and serum), pleural effusions, saliva, cerebrospinal fluid and semen (39). They have the capacity to carry proteins and nucleic acids (including DNA and RNA) from one cell to another. Exosomes are proposed to be major intermediaries of cell-cell communication in cancer and other disease and normal states (40).
Exosomes have a bilayer lipid membrane that protects them from degradation by ribonucleases and extreme pH, providing stability for nucleic acids (particularly miRNA) and allowing them to circulate for a longer period of time, when compared to the instability of circulating cell-free miRNAs (41).
All these characteristics make exosomes potential biomarkers for early diagnosis of cancer. Nonetheless, its isolation can be challenging. Different techniques have been described. The physical ones are based on molecular sizes and densities. Isolation by differential centrifugation is based on density of the molecules and allows separation of blood cellular components with different speeds of centrifugation, obtaining the exosomes at the final stage of ultracentrifugation. Ultrafiltration is another physical technique based on the size of the molecules, in which exosomes are isolated by trapping them into pores from a microscopic cylindrical structure. Exosomes can also be obtained by density gradient separation. Other non-physical, more pure methods described are polymeric-based precipitation and by immunoaffinity; the latter uses specific antibodies coated with beads, obtaining very pure exosome samples (42).
Exosomes have been shown to play a pivotal role in cancer biology, including carcinogenesis, cancer progression, invasion and metastasis. They can transfer mRNA, miRNA and proteins, promoting tumorigenesis (43). Exosomes that come from cancer cells have the capacity to transfer oncogenic factors, inducing target cells to undergo malignant transformation. One example is the transference of EGFR to vascular endothelial cells, allowing them to secrete vascular endothelial growth factor (VEGF) (44).
Exosomal miR-23a promotes lung cancer progression by 2 different mechanisms: suppressing the activity of propyl hydroxylase, increasing the accumulation of hypoxia-inducible factor-1α (part of HIF-1, a dimeric protein complex, which allows for survival and proliferation of cancerous cells due to its angiogenic properties). Exosomal miR-23a also decreases the action of tight junction proteins, facilitating transendothelial passage of malignant cells (45).
There is also a described role of exosomes in tumor metastasis. They help activate the EMT, increasing the capacity of tumor cells to circulate in the blood to distant places (46). Exosomes present in serum from patients with metastatic lung cancer can also induce expression of vimentin in other non-cancerous cells, stimulating EMT even more and increasing the potential to metastasize (47).
Nevertheless, exosomes don’t only play a role in carcinogenesis as mentioned above. They also have multiple surface proteins, making them potentially good biomarkers for early diagnosis of lung cancer. Different exosomal proteins and miRNA have been studied in lung cancer for different clinical applications, including diagnosis, staging and prognosis. Some proteins on the membrane surface of exosomes have been suggested as potential exosomal tumor markers, including CD91, CD317 and EGFR (48).
Some studies have compared exosomal miRNA from healthy individual with those from lung cancer patients in different stages and conditions, finding significant differences between those groups. Therefore, miRNA is thought to have clinical potential as tools for early diagnosis, tumor profiling, response to treatment, markers of cancer recurrence and predictors of overall survival in lung cancer. Isolation of certain miRNA has also been associated to increased ability to metastasize. Table 2 shows some of the exosomal markers (miRNA and membrane proteins) found to have potential clinical relevance. Adapted from (49) and (50).

Full table
Exosomes have multiple characteristics that make them potential drug delivery systems in the treatment of cancer. They are widely produced and secreted by almost all cell types in the human body, are able to cross membranes easily, can transport very important cellular material such as proteins, mRNA, miRNA and DNA and deliver it to target cells and are not considered toxic.
TEP
Platelets are the second most common blood cell. Its main known function is hemostasis; however, they can play other roles in different disease processes. They have the capacity to adapt to different environments and to undergo modifications, this can also happen after being in contact with lung cancer cells. Once platelets interact with tumor cells, they can become “educated”. This process is primarily driven by transference of cancer-associated biomolecules into the platelets, resulting in the creation of TEP (51).
TEP have potential clinical implications in lung cancer, helping in its diagnosis and allowing treatment monitoring. By release of their factors into tumor cells and sequestration of proteins and RNAs from cancer cells, platelets can become a key factor in tumor progression and dissemination, creation of a cancer-favorable environment by induction of angiogenesis and even protection of CTCs (52).
Platelets are not nucleated cells, but they contain a large amount of genetic material, including multiple RNA types, including miRNA, pre-mRNA, mRNA, circular RNA (cirRNA), as well as mitochondrial DNA. RNA content is higher in younger platelets compared to older platelets (53). Exposure of platelets to tumor cells induces splicing of the platelet’s RNA, allowing platelets to exchange nucleic acids and proteins with the tumor cells. Some of these proteins have functions that can benefit cancer cells in different stages of their life (54).
Platelets induce EMT that as was previously mentioned, increases the capacity of tumor cells to go into the bloodstream and invade distant tissues. Once in the bloodstream, platelets have the capacity to protect cancer cells, contributing to metastasis. Once platelets establish in the area of metastasis, they release different growth and proangiogenic factors such as VEGF, PDGF and bFG, creating an ideal environment for the proliferation of the tumor cells (55).
Platelet RNA machinery can rapidly respond to stimuli from cancer, different tumors can provoke specific mRNA splicing into the platelets, allowing identification of different type of tumors just by isolation of circulating platelets from peripheral blood. This can help distinguishing the origin of the primary tumor in up to 71% accuracy. For example, uptake of NSCLC biomarkers in platelets has been documented; these biomarkers can be used for diagnosis of NSCLC (56). Further research is needed in order to include TEP into the diagnostic arsenal for lung cancer. Due to its capacity to exchange intracellular material with tumor cells, its application as therapeutic delivery vehicle needs to be further investigated.
Circulating RNAs
Circulating RNAs are important biomarkers for liquid biopsy along with ctDNAs, CTCs, TEPs and exosomes. Stevens et al. in 1996 first described isolation of tyrosinase mRNA from peripheral blood of melanoma patients which also correlated with stage of melanoma. Most common circulating RNA in blood is microRNA (miRNA) which along with ctDNA can give a more comprehensive picture of tumor specific genetic mutations. Micro RNAs are small regulatory RNAs that can be dysregulated in cancer, they are stable and protected from endogenous RNAse activity. miRNAs can be transported to recipient cells by high density lipoproteins, exosomes and TEPs. Circulating miRNAs can be quantified by quantitative PCR (qPCR) including TaqMan PCR, NGS or hybridization technology (HYB). There are many different miRNAs which were identified to be connected with NSCLC diagnosis and prognostication. In one study a signature of four miRNAs (miR-486, miR-1, miR-30d and miR-499) were shown to be a non-invasive predictor for overall survival of NSCLC.
In 2009 Rabinowits et al. showed high concentration of circulating exosomes and miRNA in lung adenocarcinoma patients compared to controls but there was no direct correlation between concentration and stages of lung adenocarcinoma. Shen et al. by using four miRNAs (miR-126, miR-21, miR-210 and miR-486-5p) was able to detect NSCLC with 86.2% sensitivity and 96.5% specificity, including stage I NSCLC with 73.3% sensitivity and 96.5% specificity. There is also some data using circulating miRNA signatures with LDCT to increase specificity and to monitor disease recurrence in LDCT detected lung cancer patients.
Clinical applications
In current clinical practice, liquid biopsies (cfDNA/ctDNA) are not recommended to be used in lieu of tissue diagnosis; however, it is considered in certain circumstances on initial molecular diagnosis and progression during targeted therapy (57).
Treatment naïve patients considered for molecular testing in tissue and/or ctDNA are those with non-squamous NSCLC in advanced or metastatic stage, squamous NSCLC if they are never or light smokers and younger age, or if there is a non-squamous component in the tissue sample which have a higher probability to have a driver mutation (58). Table 2 summarizes relevant clinical applications of liquid biopsies.
The actionable mutations with FDA approved therapies include: Epithelial Growth Factor Receptor (EGFR) activating mutations, Anaplastic Lymphoma Kinase (ALK) translocations, BRAF V600E mutations, ROS1 gene fusions, Neurotropic Tyrosine Kinase gene (NTRK) fusions. Human Epidermal Receptor 2 (HER2) amplifications, MNNG-HOS transforming (MET) gene amplifications, MET Exon 14 skipping mutation and Rearranged during Transfection (RET) gene fusions are other mutations that have targeted therapies available however, have not yet received FDA approval (57).
Molecular testing by ctDNA can benefit patients who are medically unfit for invasive procedures if initial tissue is not enough or unavailable for molecular testing, when the procedure is too risky due to location of the suspicious lesions, if there is insufficient material for molecular testing due to the type of sample (i.e., bone biopsy) and if patients refuse re-biopsy.
Other theoretical advantages of ctDNA is that the material analyzed comes from multiple disease sites and can reflect inter and intratumoral heterogeneity.
There are different analytical methods to identify molecular alterations with ctDNA with a narrow or broad approach. A narrow approach targets alteration in relatively small regions of DNA. PCR based methods belong to this narrow approach of testing. NGS is a broad untargeted method to interrogate larger regions of DNA and multiple genes (58).
ctDNA and EGFR mutations
ctDNA is highly specific from 80 to 95% but sensitivity varies from 60% to 85%. A negative result from ctDNA should not definitely exclude the potential existence of EGFR driver mutations especially when done with PCR based methodologies since these methods have a lower sensitivity. In this scenario, the IASLC recommends NGS based ctDNA testing and/or consider obtaining a new tissue sample for repeated molecular analysis. A positive finding with PCR based methods of an actionable mutation in ctDNA is enough evidence to start targeted treatments (58).
During the screening period for the AURA extension (NCT01802632) and AURA2 (NCT02094261) study (phase 2), patients’ plasma was collected to test for EGFR T790M resistant mutation with real-time PCR in addition to tissue. In these studies, patients were treated with the third generation EGFR tyrosine kinase inhibitor (TKI) osimertinib after progressing to first- and second-generation EGFR TKI (erlotinib, gefitinib and afatinib) (59). This assay detected the EGFR T790M mutation in 61% of tumor tissue T790M mutation positive patients. The likelihood of patients having a positive ctDNA was higher in the subset of cases with extra thoracic disease versus those with only intrathoracic disease, which is probably explained by a higher tumor burden and tumor shedding into the bloodstream (60). However, irrespective of the method how EGFR T790M mutation was identified in these studies, response rate to osimertinib was the same. Resistance to first- and second-generation EGFR TKIs is mediated by T790M mutation in 60% of the cases (61). EGFR T790M mutation resistance mechanism rates are starting to decline as osimertinib has been approved in the first line setting for patients with EGFR activating mutations based on the FLAURA 3 study (62). Molecular alterations other than T790M mutations have arisen as resistance mechanism to first line osimertinib identified in plasma of patients from the FLAURA3 study with NGS. Such alterations included MET amplification (15%), EGFR C797S mutation (7%), HER2-amplification, PIK3CA and RAS mutations (2–7%). The identification of these molecular alterations by NGS instead of the limited narrow targeted mutations identify by PCR based methods, could potentially be beneficial for patients in term of clinical trial enrollment or expanded access programs.
ctDNA and ALK rearrangements
ALK (anaplastic lymphoma kinase) mutations have been traditionally identified in tissue biopsies of lung cancer adenocarcinoma specimens by fluorescence in situ hybridization (FISH) or immunohistochemistry (IHC). There is scant data on the use of ctDNA in the identification of ALK rearrangements in treatment naïve patients but certainly, there have been case reports in which ALK rearrangements were identified only in blood from patients who subsequently attained responses to ALK TKIs (63) After initiation of targeted therapy with TKIs, acquired resistance is universal and it can be detected in ctDNA with ddPCR and NGS. In a study of 20 patients with advanced ALK-positive NSCLC who progressed during crizotinib treatment; ctDNA was analyzed using ddPCR for several ALK resistant mutations (p.L1196M, p.G1269A, and p.F1174L) and Kirsten rat sarcoma (KRAS) (codons 12 and 13) mutations. In this report, five patients had ALK secondary mutations identified, one patient had two ALK mutations, and 10 patients had KRAS mutations. ddPCR sensitivity and specificity has not been investigated in the past. Most reports of ctDNA testing come from NGS modalities. The IASLC suggest that checking for ALK resistance mutations is a valuable tool for next generation TKI selection, yet not required in clinical practice to switch patients to a different ALK TKI. If testing is to be done and not possible to re-biopsy the progression sites, NGS based technology is preferred to assess not only ALK resistance mechanism, but also other molecular alterations that can make patients eligible to certain clinical trials or expanded access drug programs (58).
Liquid biopsies and other oncogenic drivers
There are other oncogenic drivers that are targetable for which existing drugs are available. National Comprehensive Cancer Network (NCCN) guidelines recommend to check for ROS1 fusions and BRAFV600 mutations in tissue samples given evidence of effective targeted therapies such as crizotinib and dabrafenib/trametinib combinations respectively. Less data exists regarding the utility of ctDNA for the detection of these targetable mutations but several NGS panels include these molecular alterations which if identified, should prompt consideration of targeted therapies. A report of NGS based liquid biopsy methodology was used in a study to identify ALK/ROS1 mutations in blood. A total of 7 patients had ROS1 fusions which were previously verified with FISH or NGS in tissue. Six of them were treatment naïve and the fusions were identified in ctDNA with a 100% of sensitivity for ROS1.
BRAF mutations have been also been identified at the time of progression in liquid biopsies using amplicon based NGS technology in patients with advanced NSCLC with BRAF mutations after BRAF ± MEK inhibitors treatment. Aggarwal et al. also reported two cases with driver BRAFV600E mutation on liquid biopsy (NGS), both of which achieved partial responses with BRAF inhibitors plus MEK inhibitors combination in one of the two cases (64).
RET-KIF5B fusions have also been identified in patients with negative molecular testing in tissue samples with hybrid-capture based NGS. There are potentially effective targeted drugs for these mutations such as vantedanib and cabozantinib, both of which represent the most heavily studied multikinase inhibitors in RET mutant NSCLC with response rates between 20% to 50% in largely pretreated patients (65-67). Other more effective drugs are currently being investigated such as BLU-667 and LOXO-292, both of which have demonstrated objective response rates of 50% and 74% in patients with RET-fusion positive NSCLC (68). In light of these promising therapies for RET-fusion positive NSCLC, identification of this molecular alterations by liquid biopsy and tissue should trigger consideration of utilizing targeted therapies as therapeutic strategies to manage these subsets of patients.
Aggarwal et al. reported four cases with MET exon 14 skipping mutations identified in ctDNA with NGS technology. These patients received crizotinib therapy achieving partial responses in one case, stable disease in two cases and one patient had progressive disease (64).
Liquid biopsies utility in the immuno-oncology era
Programmed cell death protein 1 (PD-1) and PD-L1 checkpoint inhibitors have been largely incorporated in the treatment of NSCLC in the first line and upon progression in patients with metastatic NSCLC with or without chemotherapy. Tumor mutation burden (TMB) has been studied in several clinical trials and is emerging as a surrogate for overall neoantigen load, which has been correlated to higher responses to immune check point inhibitors (69). TMB has been traditionally checked in tissue samples. Gandara et al. reported the performance of a blood-based assay to measure TMB in plasma using hybridization-capture methodology NGS targeting 1.1 Mb of genomic coding sequence. Tissue-based analysis of TMB was obtained from a subset of POPLAR and OAK samples, and blood TMB results were also checked from pre-treatment plasma from the same cohort of patients. A positive correlation between the blood and tissue TMB was detected with a Spearman rank correlation 0.64 [95% confidence interval (CI): 0.56–0.71]. Blood TMB was a predictive biomarker for progression free survival in patients receiving atezolizumab monotherapy in NSCLC when values were above of 16 mutations per megabase as a clinically meaningful and technically robust cut-point (70).
Efforts to identify PD-L1 scores in blood samples as a biomarker to predict response to checkpoint inhibitors is being studied as well, mostly in circulating cancer tumor cells. To date, this analysis has not yet been utilized in clinical practice and immunotherapy remains to be used based on PD-L1 tumor proportion scores on tissue samples (71).
Conclusions
Liquid lung biopsy not only provides important genomic information of tumors that will help with individualized treatment strategies, but it has also evolved as a promising tool for early detection of lung cancer and disease monitoring to help further with management decisions in selected patients. There are several biomarkers from which these somatic alterations are obtained; the more studied and used ones being tests analyzing ctDNA. Even though other biomarkers are still the subject of ongoing research, understanding their role in carcinogenesis and cancer behavior is crucial to develop further testing methods. It is worth mentioning that in the era of evolving minimally invasive diagnostic pulmonary medicine, tumor tissue acquisition is more feasible and quicker than in the past with a low risk profile compared to surgical sampling, however, with the potential caveat of missing the whole make up (intratumoral and intertumoral heterogeneity) with a single-site biopsy. As liquid lung biopsy methods become more standardized, tissue sampling remains gold standard. Furthermore, the highest sensitivity of commercially available testing platforms is about 85%, posing a risk for false negative results, especially in those treatment-naïve patients. Finally, it is worth mentioning that liquid biopsies are obtained from a simple blood draw making it an easily reproducible, low risk and perhaps with a low overall cost compared to other methods.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Boloker G, Wang C, Zhang J. Updated statistics of lung and bronchus cancer in United States (2018). J Thorac Dis 2018;10:1158-61. [Crossref] [PubMed]
- Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2018;4:1553-68. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Patz EF Jr, Campa MJ, Gottlin EB, et al. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol 2007;25:5578-83. [Crossref] [PubMed]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- De Koning H, Van Der Aalst C, Ten Haaf K, et al. Effects of volume CT lung cancer screening: Mortality results of the NELSON randomized-controlled population based trial. J Thorac Oncol 2018;13:S185. [Crossref]
- Tofts RP, Lee PM, Sung AW. Interventional pulmonology approaches in the diagnosis and treatment of early stage non small cell lung cancer. Transl Lung Cancer Res 2013;2:316-31. [PubMed]
- He C, Liu M, Zhou C, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 2009;125:2393-9. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Shukuya T, Patel S, Shane-Carson K, et al. Lung Cancer Patients with Germline Mutations Detected by Next-Generation Sequencing and/or Liquid Biopsy. J Thorac Oncol 2018;13:e17-9. [Crossref] [PubMed]
- Bauml J, Levy B. Clonal Hematopoiesis: A New Layer in the Liquid Biopsy Story in Lung Cancer. Clin Cancer Res 2018;24:4352-4. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Lim M, Kim CJ, Sunkara V, et al. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and ctDNA). Micromachines (Basel) 2018.9. [PubMed]
- Bracht JWP, Mayo-de-Las-Casas C, Berenguer J, et al. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr Oncol Rep 2018;20:70. [Crossref] [PubMed]
- Schmidt B, Liebenberg V, Dietrich D, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 2010;10:600. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Santarpia M, Liguori A, D'Aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis 2018;10:S882-97. [Crossref] [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013;81:397-403. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016;63:246-53. [PubMed]
- Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A Novel Epigenetic Signature for Early Diagnosis in Lung Cancer. Clin Cancer Res 2016;22:3361-71. [Crossref] [PubMed]
- O'Flaherty L, Wikman H, Pantel K. Biology and clinical significance of circulating tumor cell subpopulations in lung cancer. Transl Lung Cancer Res 2017;6:431-43. [Crossref] [PubMed]
- Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: What we know and what we expect Int J Oncol 2016;49:2206-16. (Review). [Crossref] [PubMed]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442-54. [Crossref] [PubMed]
- Bidard FC, Proudhon C, Pierga JY. Circulating tumor cells in breast cancer. Mol Oncol 2016;10:418-30. [Crossref] [PubMed]
- Zhang T, Armstrong AJ. Clinical Utility of Circulating Tumor Cells in Advanced Prostate Cancer. Curr Oncol Rep 2016;18:3. [Crossref] [PubMed]
- Hinz S, Bockhorst J, Roder C, et al. Disseminated tumor cells in the bone marrow negatively influence survival after resection of colorectal liver metastases. Ann Surg Oncol 2012;19:2539-46. [Crossref] [PubMed]
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006;6:449-58. [Crossref] [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Alix-Panabieres C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95:133-42. [Crossref] [PubMed]
- Mahmood MQ, Ward C, Muller HK, et al. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): a mutual association with airway disease. Med Oncol 2017;34:45. [Crossref] [PubMed]
- Babayan A, Alawi M, Gormley M, et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 2016;8:56066-80. [PubMed]
- Zheng H, Zhan Y, Liu S, et al. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res 2018;37:226. [Crossref] [PubMed]
- van der Pol E, Boing AN, Harrison P, et al. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 2012;64:676-705. [Crossref] [PubMed]
- Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn 2013;15:827-34. [Crossref] [PubMed]
- Vanni I, Alama A, Grossi F, et al. Exosomes: a new horizon in lung cancer. Drug Discov Today 2017;22:927-36. [Crossref] [PubMed]
- Kharaziha P, Ceder S, Li Q, et al. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta 2012;1826:103-11. [PubMed]
- Abdouh M, Hamam D, Gao ZH, et al. Exosomes isolated from cancer patients' sera transfer malignant traits and confer the same phenotype of primary tumors to oncosuppressor-mutated cells. J Exp Clin Cancer Res 2017;36:113. [Crossref] [PubMed]
- Wu H, Zhou J, Mei S, et al. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med 2017;21:1228-36. [Crossref] [PubMed]
- Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431-7. [Crossref] [PubMed]
- Zomer A, Maynard C, Verweij FJ, et al. In Vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015;161:1046-57. [Crossref] [PubMed]
- Yamashita T, Kamada H, Kanasaki S, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013;68:969-73. [PubMed]
- Zhou L, Lv T, Zhang Q, et al. The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett 2017;407:84-92. [Crossref] [PubMed]
- Liu S, Zhan Y, Luo J, et al. Roles of exosomes in the carcinogenesis and clinical therapy of non-small cell lung cancer. Biomed Pharmacother 2019;111:338-46. [Crossref] [PubMed]
- McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717-27. [Crossref] [PubMed]
- Joosse SA, Pantel K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. Cancer Cell 2015;28:552-4. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Alhasan AA, Izuogu OG, Al-Balool HH, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016;127:e1-11. [Crossref] [PubMed]
- Klement GL, Yip TT, Cassiola F, et al. Platelets actively sequester angiogenesis regulators. Blood 2009;113:2835-42. [Crossref] [PubMed]
- Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-21. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758-70. [Crossref] [PubMed]
- Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol 2019;5:173-80. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Gautschi O, Milia J, Filleron T, et al. Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J Clin Oncol 2017;35:1403-10. [Crossref] [PubMed]
- Suzuki M, Makinoshima H, Matsumoto S, et al. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci 2013;104:896-903. [Crossref] [PubMed]
- Subbiah V, Gainor JF, Rahal R, et al. Precision Targeted Therapy with BLU-667 for RET-Driven Cancers. Cancer Discov 2018;8:836-49. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Dhar M, Wong J, Che J, et al. Evaluation of PD-L1 expression on vortex-isolated circulating tumor cells in metastatic lung cancer. Sci Rep 2018;8:2592. [Crossref] [PubMed]


